Pool-GWAS on reproductive dormancy in Drosophila simulans suggests a polygenic architecture
- PMID: 35137042
- PMCID: PMC8895979
- DOI: 10.1093/g3journal/jkac027
Pool-GWAS on reproductive dormancy in Drosophila simulans suggests a polygenic architecture
Abstract
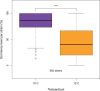
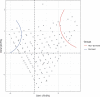
The genetic basis of adaptation to different environments has been of long-standing interest to evolutionary biologists. Dormancy is a well-studied adaptation to facilitate overwintering. In Drosophila melanogaster, a moderate number of genes with large effects have been described, which suggests a simple genetic basis of dormancy. On the other hand, genome-wide scans for dormancy suggest a polygenic architecture in insects. In D. melanogaster, the analysis of the genetic architecture of dormancy is complicated by the presence of cosmopolitan inversions. Here, we performed a genome-wide scan to characterize the genetic basis of this ecologically extremely important trait in the sibling species of D. melanogaster, D. simulans that lacks cosmopolitan inversions. We performed Pool-GWAS in a South African D. simulans population for dormancy incidence at 2 temperature regimes (10 and 12°C, LD 10:14). We identified several genes with SNPs that showed a significant association with dormancy (P-value < 1e-13), but the overall modest response suggests that dormancy is a polygenic trait with many loci of small effect. Our results shed light on controversies on reproductive dormancy in Drosophila and have important implications for the characterization of the genetic basis of this trait.
Keywords: Drosophila; adaptation; dormancy; genetic architecture.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Figures


Similar articles
-
Redefining reproductive dormancy in Drosophila as a general stress response to cold temperatures.J Insect Physiol. 2018 May-Jun;107:175-185. doi: 10.1016/j.jinsphys.2018.04.006. Epub 2018 Apr 9. J Insect Physiol. 2018. PMID: 29649483
-
Genetic redundancy fuels polygenic adaptation in Drosophila.PLoS Biol. 2019 Feb 4;17(2):e3000128. doi: 10.1371/journal.pbio.3000128. eCollection 2019 Feb. PLoS Biol. 2019. PMID: 30716062 Free PMC article.
-
Drosophila simulans: A Species with Improved Resolution in Evolve and Resequence Studies.G3 (Bethesda). 2017 Jul 5;7(7):2337-2343. doi: 10.1534/g3.117.043349. G3 (Bethesda). 2017. PMID: 28546383 Free PMC article.
-
Comparative analysis of morphological traits among Drosophila melanogaster and D. simulans: genetic variability, clines and phenotypic plasticity.Genetica. 2004 Mar;120(1-3):165-79. doi: 10.1023/b:gene.0000017639.62427.8b. Genetica. 2004. PMID: 15088656 Review.
-
Historicity and the population genetics of Drosophila melanogaster and D. simulans.Genetica. 2004 Mar;120(1-3):61-70. doi: 10.1023/b:gene.0000017630.69020.32. Genetica. 2004. PMID: 15088647 Review.
Cited by
-
Extreme trait GWAS (Et-GWAS): Unraveling rare variants in the 3,000 rice genome.Life Sci Alliance. 2023 Dec 26;7(3):e202302352. doi: 10.26508/lsa.202302352. Print 2024 Mar. Life Sci Alliance. 2023. PMID: 38148113 Free PMC article.
References
-
- Anscombe FJ. On estimating binomial response relations. Biometrika. 1956;43(3–4):461–464. doi:10.1093/biomet/43.3-4.461. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
