Clinical and molecular assessment of an onco-immune signature with prognostic significance in patients with colorectal cancer
- PMID: 35137551
- PMCID: PMC8921909
- DOI: 10.1002/cam4.4568
Clinical and molecular assessment of an onco-immune signature with prognostic significance in patients with colorectal cancer
Abstract
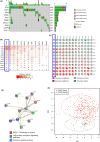
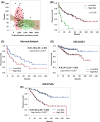
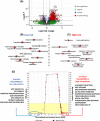
Understanding the complex tumor microenvironment is key to the development of personalized therapies for the treatment of cancer including colorectal cancer (CRC). In the past decade, significant advances in the field of immunotherapy have changed the paradigm of cancer treatment. Despite significant improvements, tumor heterogeneity and lack of appropriate classification tools for CRC have prevented accurate risk stratification and identification of a wider patient population that may potentially benefit from targeted therapies. To identify novel signatures for accurate prognostication of CRC, we quantified gene expression of 12 immune-related genes using a medium-throughput NanoString quantification platform in 93 CRC patients. Multivariate prognostic analysis identified a combined four-gene prognostic signature (TGFB1, PTK2, RORC, and SOCS1) (HR: 1.76, 95% CI: 1.05-2.95, *p < 0.02). The survival trend was captured in an independent gene expression data set: GSE17536 (177 patients; HR: 3.31, 95% CI: 1.99-5.55, *p < 0.01) and GSE14333 (226 patients; HR: 2.47, 95% CI: 1.35-4.53, *p < 0.01). Further, gene set enrichment analysis of the TCGA data set associated higher prognostic scores with epithelial-mesenchymal transition (EMT) and inflammatory pathways. Comparatively, a lower prognostic score was correlated with oxidative phosphorylation and MYC and E2F targets. Analysis of immune parameters identified infiltration of T-reg cells, CD8+ T cells, M2 macrophages, and B cells in high-risk patient groups along with upregulation of immune exhaustion genes. This molecular study has identified a novel prognostic gene signature with clinical utility in CRC. Therefore, along with prognostic features, characterization of immune cell infiltrates and immunosuppression provides actionable information that should be considered while employing personalized medicine.
Keywords: colon; colorectal cancer; gene signature; immune; personalized medicine; preventive; prognostic genes.
© 2022 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Nordlinger B, Sorbye H, Glimelius B, et al.; EORTC Gastro‐Intestinal Tract Cancer Group, Cancer Research UK, Arbeitsgruppe Lebermetastasen und–tumoren in der Chirurgischen Arbeitsgemeinschaft Onkologie (ALM‐CAO), Australasian Gastro‐Intestinal Trials Group (AGITG), Fédération Francophone de Cancérologie Digestive (FFCD) . Perioperative FOLFOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): long‐term results of a randomised, controlled, phase 3 trial. Lancet Oncol. 2013;14(12):1208–1215. doi:10.1016/S1470-2045(13)70447-9 - DOI - PubMed
-
- Turin I, Delfanti S, Ferulli F, et al. In vitro killing of colorectal carcinoma cells by autologous activated NK cells is boosted by anti‐epidermal growth factor receptor‐induced ADCC regardless of RAS mutation status. J Immunother. 2018;41(4):190‐200. doi:10.1097/CJI.0000000000000205 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

