Doramectin inhibits glioblastoma cell survival via regulation of autophagy in vitro and in vivo
- PMID: 35137919
- PMCID: PMC8857930
- DOI: 10.3892/ijo.2022.5319
Doramectin inhibits glioblastoma cell survival via regulation of autophagy in vitro and in vivo
Abstract
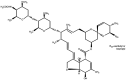
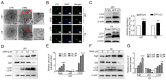
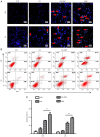
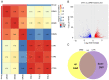
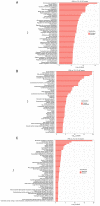
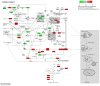
Glioblastoma (GBM) is one of the most widespread and lethal types of cancer. However, there are currently no drugs or therapeutic strategies that can completely cure GBM. Doramectin (DRM) has a broad range of activities against endoparasites and ectoparasites, and is extensively used in livestock. In the present study, the effect of DRM on the induction of autophagy in U87 and C6 GBM and glioma cell lines, as well as the mechanism of autophagy, were examined. First, transmission electron microscopy, plasmid transfection and western blot analysis demonstrated that DRM could induce autophagy in U87 and C6 cells in vitro. Next, MTT and colony formation assays revealed that DRM‑induced autophagy prevented U87 and C6 cell viability and colony formation ratio. In addition, DRM‑induced autophagy promoted U87 and C6 cell apoptosis, as indicated by DAPI analysis and flow cytometry. Furthermore, transcriptome analysis demonstrated that DRM modulated a number of genes and pathways involved in autophagy. In a nude mouse xenograft model, immunohistochemical staining and the TUNEL assay demonstrated that the effect of DRM on the tumor was consistent with that in vivo. These data indicated that DRM induced autophagy mainly by blocking the PI3K/AKT/mTOR signaling pathway in GBM cells. DRM‑induced autophagy promoted the inhibition of GBM cell proliferation and apoptosis in vitro and in vivo. The present study suggested that DRM may be an effective drug for the treatment of GBM.
Keywords: PI3K/AKT/mTOR; apoptosis; autophagy; glioblastoma; transcriptomic analysis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
