Construction and characterization of two SARS-CoV-2 minigenome replicon systems
- PMID: 35137972
- PMCID: PMC9088700
- DOI: 10.1002/jmv.27650
Construction and characterization of two SARS-CoV-2 minigenome replicon systems
Abstract
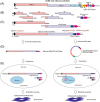
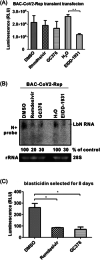
The ongoing COVID-19 pandemic severely impacts global public health and economies. To facilitate research on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virology and antiviral discovery, a noninfectious viral replicon system operating under biosafety level 2 containment is warranted. We report herein the construction and characterization of two SARS-CoV-2 minigenome replicon systems. First, we constructed the IVT-CoV2-Rep complementary DNA template to generate a replicon messenger RNA (mRNA) with nanoluciferase (NLuc) reporter via in vitro transcription (IVT). The replicon mRNA transfection assay demonstrated a rapid and transient replication of IVT-CoV2-Rep in a variety of cell lines, which could be completely abolished by known SARS-CoV-2 replication inhibitors. Our data also suggest that the transient phenotype of IVT-CoV2-Rep is not due to host innate antiviral responses. In addition, we have developed a DNA-launched replicon BAC-CoV2-Rep, which supports the in-cell transcription of a replicon mRNA as initial replication template. The BAC-CoV2-Rep transient transfection system exhibited a much stronger and longer replicon signal compared to the IVT-CoV2-Rep version. We also found that a portion of the NLuc reporter signal was derived from the spliced BAC-CoV2-Rep mRNA and was resistant to antiviral treatment, especially during the early phase after transfection. In summary, the established SARS-CoV-2 transient replicon systems are suitable for basic and antiviral research, and hold promise for stable replicon cell line development with further optimization.
Keywords: SARS coronavirus; antiviral agents; cellular effect; disease control; immune responses; innate immunity; mRNA/splicing; virus classification.
© 2022 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

