Keap1 recognizes EIAV early accessory protein Rev to promote antiviral defense
- PMID: 35139135
- PMCID: PMC8863222
- DOI: 10.1371/journal.ppat.1009986
Keap1 recognizes EIAV early accessory protein Rev to promote antiviral defense
Abstract
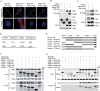
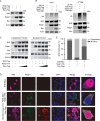
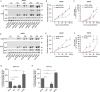
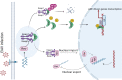
The Nrf2/Keap1 axis plays a complex role in viral susceptibility, virus-associated inflammation and immune regulation in host cells. However, whether or how the Nrf2/Keap1 axis is involved in the interactions between equine lentiviruses and their hosts remains unclear. Here, we demonstrate that the Nrf2/Keap1 axis was activated during EIAV infection. Mechanistically, EIAV-Rev competitively binds to Keap1 and releases Nrf2 from Keap1-mediated repression, leading to the accumulation of Nrf2 in the nucleus and promoting Nrf2 responsive genes transcription. Subsequently, we demonstrated that the Nrf2/Keap1 axis represses EIAV replication via two independent molecular mechanisms: directly increasing antioxidant enzymes to promote effective cellular resistance against EIAV infection, and repression of Rev-mediated RNA transport through direct interaction between Keap1 and Rev. Together, these data suggest that activation of the Nrf2/Keap1 axis mediates a passive defensive response to combat EIAV infection. The Nrf2/Keap1 axis could be a potential target for developing strategies for combating EIAV infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Identification of a Novel Post-transcriptional Transactivator from the Equine Infectious Anemia Virus.J Virol. 2022 Dec 21;96(24):e0121022. doi: 10.1128/jvi.01210-22. Epub 2022 Nov 30. J Virol. 2022. PMID: 36448796 Free PMC article.
-
NF-κB and Keap1 Interaction Represses Nrf2-Mediated Antioxidant Response in Rabbit Hemorrhagic Disease Virus Infection.J Virol. 2020 May 4;94(10):e00016-20. doi: 10.1128/JVI.00016-20. Print 2020 May 4. J Virol. 2020. PMID: 32161178 Free PMC article.
-
Caffeic acid inhibits HCV replication via induction of IFNα antiviral response through p62-mediated Keap1/Nrf2 signaling pathway.Antiviral Res. 2018 Jun;154:166-173. doi: 10.1016/j.antiviral.2018.04.008. Epub 2018 Apr 12. Antiviral Res. 2018. PMID: 29656059
-
Nrf2/Keap1/ARE signaling: Towards specific regulation.Life Sci. 2022 Feb 15;291:120111. doi: 10.1016/j.lfs.2021.120111. Epub 2021 Oct 31. Life Sci. 2022. PMID: 34732330 Free PMC article. Review.
-
Management of COVID-19-induced cytokine storm by Keap1-Nrf2 system: a review.Inflammopharmacology. 2021 Oct;29(5):1347-1355. doi: 10.1007/s10787-021-00860-5. Epub 2021 Aug 9. Inflammopharmacology. 2021. PMID: 34373972 Free PMC article. Review.
Cited by
-
A Novel, Fully Spliced, Accessory Gene in Equine Lentivirus with Distinct Rev-Responsive Element.J Virol. 2022 Sep 28;96(18):e0098622. doi: 10.1128/jvi.00986-22. Epub 2022 Sep 7. J Virol. 2022. PMID: 36069548 Free PMC article.
-
Equine Infectious Anemia Virus Cellular Partners Along the Viral Cycle.Viruses. 2024 Dec 24;17(1):5. doi: 10.3390/v17010005. Viruses. 2024. PMID: 39861793 Free PMC article. Review.
-
Identification of a Novel Post-transcriptional Transactivator from the Equine Infectious Anemia Virus.J Virol. 2022 Dec 21;96(24):e0121022. doi: 10.1128/jvi.01210-22. Epub 2022 Nov 30. J Virol. 2022. PMID: 36448796 Free PMC article.
-
Development and evaluation of a blocking ELISA for serological diagnosis of equine infectious anemia.Appl Microbiol Biotechnol. 2023 May;107(10):3305-3317. doi: 10.1007/s00253-023-12504-5. Epub 2023 Apr 11. Appl Microbiol Biotechnol. 2023. PMID: 37039847
-
Hesperidin ameliorates H2O2-induced bovine mammary epithelial cell oxidative stress via the Nrf2 signaling pathway.J Anim Sci Biotechnol. 2024 Apr 9;15(1):57. doi: 10.1186/s40104-024-01012-9. J Anim Sci Biotechnol. 2024. PMID: 38589950 Free PMC article.
References
-
- Haas L. [Equine infectious anemia—a review]. Berl Munch Tierarztl Wochenschr. 2014;127(7–8):297–300. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

