Pseudomonas aeruginosa modulates alginate biosynthesis and type VI secretion system in two critically ill COVID-19 patients
- PMID: 35139898
- PMCID: PMC8827185
- DOI: 10.1186/s13578-022-00748-z
Pseudomonas aeruginosa modulates alginate biosynthesis and type VI secretion system in two critically ill COVID-19 patients
Abstract
Background: COVID-19 pneumonia has caused huge impact on the health of infected patients and associated with high morbidity and mortality. Shift in the lung microbial ecology upon such viral infection often worsens the disease and increases host susceptibility to superinfections. Bacterial superinfection contributes to the aggravation of COVID-19 and poses a great challenge to clinical treatments. An in-depth investigation on superinfecting bacteria in COVID-19 patients might facilitate understanding of lung microenvironment post virus infections and superinfection mechanism.
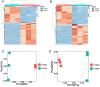
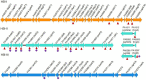
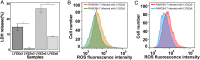
Results: We analyzed the adaptation of two pairs of P. aeruginosa strains with the same MLST type isolated from two critical COVID-19 patients by combining sequencing analysis and phenotypic assays. Both P. aeruginosa strains were found to turn on alginate biosynthesis and attenuate type VI secretion system (T6SS) during short-term colonization in the COVID-19 patients, which results in excessive biofilm formation and virulence reduction-two distinct markers for chronic infections. The macrophage cytotoxicity test and intracellular reactive oxygen species measurement confirmed that the adapted P. aeruginosa strains reduced their virulence towards host cells and are better to escape from host immune clearance than their ancestors.
Conclusion: Our study suggests that SARS-CoV-2 infection can create a lung environment that allow rapid adaptive evolution of bacterial pathogens with genetic traits suitable for chronic infections.
Keywords: Bacterial superinfection; Biofilm; COVID-19; Pseudomonas aeruginosa; Type VI Secretion System.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Grants and funding
- 2020B1515020003/Guangdong Natural Science Foundation for Distinguished Young Scholar
- JCYJ20190809144005609/Science and Technology Program of Shenzhen
- 2020A1515010586/Guangdong Basic and Applied Basic Research Foundation
- 2020B1111170014/Guangdong Province Science and Technology Project
- ZDSYS20200811144002008/Shenzhen Innovation Committee of Science and Technology
LinkOut - more resources
Full Text Sources
Miscellaneous

