Dynamic Runx1 chromatin boundaries affect gene expression in hematopoietic development
- PMID: 35140205
- PMCID: PMC8828719
- DOI: 10.1038/s41467-022-28376-8
Dynamic Runx1 chromatin boundaries affect gene expression in hematopoietic development
Abstract
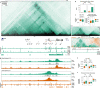
The transcription factor RUNX1 is a critical regulator of developmental hematopoiesis and is frequently disrupted in leukemia. Runx1 is a large, complex gene that is expressed from two alternative promoters under the spatiotemporal control of multiple hematopoietic enhancers. To dissect the dynamic regulation of Runx1 in hematopoietic development, we analyzed its three-dimensional chromatin conformation in mouse embryonic stem cell (ESC) differentiation cultures. Runx1 resides in a 1.1 Mb topologically associating domain (TAD) demarcated by convergent CTCF motifs. As ESCs differentiate to mesoderm, chromatin accessibility, Runx1 enhancer-promoter (E-P) interactions, and CTCF-CTCF interactions increase in the TAD, along with initiation of Runx1 expression from the P2 promoter. Differentiation to hematopoietic progenitor cells is associated with the formation of tissue-specific sub-TADs over Runx1, a shift in E-P interactions, P1 promoter demethylation, and robust expression from both Runx1 promoters. Deletion of promoter-proximal CTCF sites at the sub-TAD boundaries has no obvious effects on E-P interactions but leads to partial loss of domain structure, mildly affects gene expression, and delays hematopoietic development. Together, our analysis of gene regulation at a large multi-promoter developmental gene reveals that dynamic sub-TAD chromatin boundaries play a role in establishing TAD structure and coordinated gene expression.
© 2022. The Author(s).
Conflict of interest statement
J.R.Hu and D.J. are founders and shareholders of, and D.J.D. and R.S. are paid consultants for Nucleome Therapeutics. Other authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
- G0902418/MRC_/Medical Research Council/United Kingdom
- MR/T014067/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00029/3/MRC_/Medical Research Council/United Kingdom
- 105281/Z/14/Z/WT_/Wellcome Trust/United Kingdom
- 106130/Z/14/Z/WT_/Wellcome Trust/United Kingdom
- MR/X001210/1/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MC_UU_00016/14/MRC_/Medical Research Council/United Kingdom
- 108870/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- MC_UU_00016/2/MRC_/Medical Research Council/United Kingdom
- 203728/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- MC_UU_12009/2/MRC_/Medical Research Council/United Kingdom
- MR/N00969X/1 /MRC_/Medical Research Council/United Kingdom
- MR/K015777X/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

