Single-cell analysis of human glioma and immune cells identifies S100A4 as an immunotherapy target
- PMID: 35140215
- PMCID: PMC8828877
- DOI: 10.1038/s41467-022-28372-y
Single-cell analysis of human glioma and immune cells identifies S100A4 as an immunotherapy target
Abstract
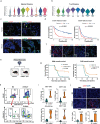
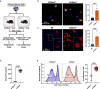
A major rate-limiting step in developing more effective immunotherapies for GBM is our inadequate understanding of the cellular complexity and the molecular heterogeneity of immune infiltrates in gliomas. Here, we report an integrated analysis of 201,986 human glioma, immune, and other stromal cells at the single cell level. In doing so, we discover extensive spatial and molecular heterogeneity in immune infiltrates. We identify molecular signatures for nine distinct myeloid cell subtypes, of which five are independent prognostic indicators of glioma patient survival. Furthermore, we identify S100A4 as a regulator of immune suppressive T and myeloid cells in GBM and demonstrate that deleting S100a4 in non-cancer cells is sufficient to reprogram the immune landscape and significantly improve survival. This study provides insights into spatial, molecular, and functional heterogeneity of glioma and glioma-associated immune cells and demonstrates the utility of this dataset for discovering therapeutic targets for this poorly immunogenic cancer.
© 2022. The Author(s).
Conflict of interest statement
K.Y. is a co-founder of EMPIRI, Inc. The remaining authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

