Translocated Legionella pneumophila small RNAs mimic eukaryotic microRNAs targeting the host immune response
- PMID: 35140216
- PMCID: PMC8828724
- DOI: 10.1038/s41467-022-28454-x
Translocated Legionella pneumophila small RNAs mimic eukaryotic microRNAs targeting the host immune response
Abstract
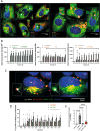
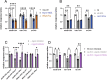
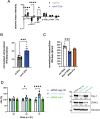
Legionella pneumophila is an intracellular bacterial pathogen that can cause a severe form of pneumonia in humans, a phenotype evolved through interactions with aquatic protozoa in the environment. Here, we show that L. pneumophila uses extracellular vesicles to translocate bacterial small RNAs (sRNAs) into host cells that act on host defence signalling pathways. The bacterial sRNA RsmY binds to the UTR of ddx58 (RIG-I encoding gene) and cRel, while tRNA-Phe binds ddx58 and irak1 collectively reducing expression of RIG-I, IRAK1 and cRel, with subsequent downregulation of IFN-β. Thus, RsmY and tRNA-Phe are bacterial trans-kingdom regulatory RNAs downregulating selected sensor and regulator proteins of the host cell innate immune response. This miRNA-like regulation of the expression of key sensors and regulators of immunity is a feature of L. pneumophila host-pathogen communication and likely represents a general mechanism employed by bacteria that interact with eukaryotic hosts.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Mondino S, et al. Legionnaires’ disease: State of the art knowledge of pathogenesis mechanisms of Legionella. Annu. Rev. Pathol. 2020;15:439–466. - PubMed
-
- Escoll P, Rolando M, Gomez-Valero L, Buchrieser C. From amoeba to macrophages: Exploring the molecular mechanisms of Legionella pneumophila Infection in both hosts. Curr. Top. Microbiol. Immunol. 2013;376:1–34. - PubMed
-
- Cazalet C, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 2004;36:1165–1173. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

