A platform for oncogenomic reporting and interpretation
- PMID: 35140225
- PMCID: PMC8828759
- DOI: 10.1038/s41467-022-28348-y
A platform for oncogenomic reporting and interpretation
Abstract
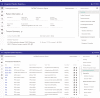
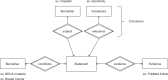
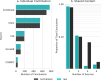
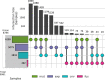
Manual interpretation of variants remains rate limiting in precision oncology. The increasing scale and complexity of molecular data generated from comprehensive sequencing of cancer samples requires advanced interpretative platforms as precision oncology expands beyond individual patients to entire populations. To address this unmet need, we introduce a Platform for Oncogenomic Reporting and Interpretation (PORI), comprising an analytic framework that facilitates the interpretation and reporting of somatic variants in cancer. PORI integrates reporting and graph knowledge base tools combined with support for manual curation at the reporting stage. PORI represents an open-source platform alternative to commercial reporting solutions suitable for comprehensive genomic data sets in precision oncology. We demonstrate the utility of PORI by matching 9,961 pan-cancer genome atlas tumours to the graph knowledge base, calculating therapeutically informative alterations, and making available reports describing select individual samples.
© 2022. The Author(s).
Conflict of interest statement
Dr. Stephen Yip has received funding as a member of scientific advisory boards of Amgen, AstraZeneca, Bayer, Norvatis, and Roche. All other authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

