Guanosine primes acute myeloid leukemia for differentiation via guanine nucleotide salvage synthesis
- PMID: 35141027
- PMCID: PMC8822274
Guanosine primes acute myeloid leukemia for differentiation via guanine nucleotide salvage synthesis
Abstract
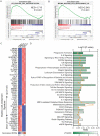
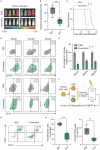
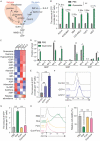
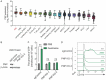
Differentiation arrest represents a distinct hallmark of acute myeloid leukemia (AML). Identification of differentiation-induction agents that are effective across various subtypes remains an unmet challenge. GTP biosynthesis is elevated in several types of cancers, considered to support uncontrolled tumor growth. Here we report that GTP overload by supplementation of guanosine, the nucleoside precursor of GTP, poises AML cells for differentiation and growth inhibition. Transcriptome profiling of guanosine-treated AML cells reveals a myeloid differentiation pattern. Importantly, the treatment compromises leukemia progression in AML xenograft models. Mechanistically, GTP overproduction requires sequential metabolic conversions executed by the purine salvage biosynthesis pathway including the involvement of purine nucleoside phosphorylase (PNP) and hypoxanthine phosphoribosyltransferase 1 (HPRT1). Taken together, our study offers novel metabolic insights tethering GTP homeostasis to myeloid differentiation and provides an experimental basis for further clinical investigations of guanosine or guanine nucleotides in the treatment of AML patients.
Keywords: Acute myeloid leukemia; HPRT1; PNP; guanosine 5’-triphosphate; myeloid differentiation.
AJCR Copyright © 2022.
Conflict of interest statement
None.
Figures

Similar articles
-
Profiles of purine biosynthesis, salvage and degradation in disks of potato (Solanum tuberosum L.) tubers.Planta. 2006 Dec;225(1):115-26. doi: 10.1007/s00425-006-0334-9. Epub 2006 Jul 15. Planta. 2006. PMID: 16845529
-
2'-deoxyguanosine toxicity for B and mature T lymphoid cell lines is mediated by guanine ribonucleotide accumulation.J Clin Invest. 1984 Nov;74(5):1640-8. doi: 10.1172/JCI111580. J Clin Invest. 1984. PMID: 6334099 Free PMC article.
-
Mechanisms of deoxyguanosine lymphotoxicity. Human thymocytes, but not peripheral blood lymphocytes accumulate deoxy-GTP in conditions simulating purine nucleoside phosphorylase deficiency.J Immunol. 1990 Jan 15;144(2):485-91. J Immunol. 1990. PMID: 2104895
-
Trophic effects of purines in neurons and glial cells.Prog Neurobiol. 1999 Dec;59(6):663-90. doi: 10.1016/s0301-0082(99)00017-9. Prog Neurobiol. 1999. PMID: 10845757 Review.
-
Older and new purine nucleoside analogs for patients with acute leukemias.Cancer Treat Rev. 2013 Dec;39(8):851-61. doi: 10.1016/j.ctrv.2013.03.006. Epub 2013 Apr 6. Cancer Treat Rev. 2013. PMID: 23566572 Review.
Cited by
-
Guanine inhibits the growth of human glioma and melanoma cell lines by interacting with GPR23.Front Pharmacol. 2022 Sep 19;13:970891. doi: 10.3389/fphar.2022.970891. eCollection 2022. Front Pharmacol. 2022. PMID: 36199684 Free PMC article.
-
Lessons from the physiological role of guanosine in neurodegeneration and cancer: Toward a multimodal mechanism of action?Purinergic Signal. 2025 Feb;21(1):133-148. doi: 10.1007/s11302-024-10033-y. Epub 2024 Jul 15. Purinergic Signal. 2025. PMID: 39004650 Free PMC article. Review.
-
Guanine nucleotide biosynthesis blockade impairs MLL complex formation and sensitizes leukemias to menin inhibition.Nat Commun. 2025 Mar 18;16(1):2641. doi: 10.1038/s41467-025-57544-9. Nat Commun. 2025. PMID: 40102405 Free PMC article.
References
-
- Talati C, Sweet K. Recently approved therapies in acute myeloid leukemia: a complex treatment landscape. Leuk Res. 2018;73:58–66. - PubMed
-
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, Gundem G, Van Loo P, Martincorena I, Ganly P, Mudie L, McLaren S, O’Meara S, Raine K, Jones DR, Teague JW, Butler AP, Greaves MF, Ganser A, Döhner K, Schlenk RF, Döhner H, Campbell PJ. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374:2209–2221. - PMC - PubMed
-
- de Thé H. Differentiation therapy revisited. Nat Rev Cancer. 2018;18:117–127. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
