Severe COVID-19 Is Associated With an Altered Upper Respiratory Tract Microbiome
- PMID: 35141167
- PMCID: PMC8819187
- DOI: 10.3389/fcimb.2021.781968
Severe COVID-19 Is Associated With an Altered Upper Respiratory Tract Microbiome
Abstract
Background: The upper respiratory tract (URT) is the portal of entry of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and SARS-CoV-2 likely interacts with the URT microbiome. However, understanding of the associations between the URT microbiome and the severity of coronavirus disease 2019 (COVID-19) is still limited.
Objective: Our primary objective was to identify URT microbiome signature/s that consistently changed over a spectrum of COVID-19 severity.
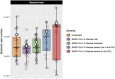
Methods: Using data from 103 adult participants from two cities in the United States, we compared the bacterial load and the URT microbiome between five groups: 20 asymptomatic SARS-CoV-2-negative participants, 27 participants with mild COVID-19, 28 participants with moderate COVID-19, 15 hospitalized patients with severe COVID-19, and 13 hospitalized patients in the ICU with very severe COVID-19.
Results: URT bacterial load, bacterial richness, and within-group microbiome composition dissimilarity consistently increased as COVID-19 severity increased, while the relative abundance of an amplicon sequence variant (ASV), Corynebacterium_unclassified.ASV0002, consistently decreased as COVID-19 severity increased.
Conclusions: We observed that the URT microbiome composition significantly changed as COVID-19 severity increased. The URT microbiome could potentially predict which patients may be more likely to progress to severe disease or be modified to decrease severity. However, further research in additional longitudinal cohorts is needed to better understand how the microbiome affects COVID-19 severity.
Keywords: COVID-19; SARS-CoV-2; microbiome; mild; moderate; severe COVID-19 outcomes; upper respiratory tract.
Copyright © 2022 Shilts, Rosas-Salazar, Strickland, Kimura, Asad, Sehanobish, Freeman, Wessinger, Gupta, Brown, Boone, Patel, Barbi, Bottalico, O’Neill, Akbar, Rajagopala, Mallal, Phillips, Turner, Jerschow and Das.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Anderson M. J. (2001). A New Method for Non-Parametric Multivariate Analysis of Variance. Austral Ecol. 26, 32–46.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

