Plasma microRNA vary in association with the progression of Alzheimer's disease
- PMID: 35141392
- PMCID: PMC8817674
- DOI: 10.1002/dad2.12251
Plasma microRNA vary in association with the progression of Alzheimer's disease
Abstract
Introduction: Early intervention in Alzheimer's disease (AD) requires the development of an easily administered test that is able to identify those at risk. Focusing on microRNA robustly detected in plasma and standardizing the analysis strategy, we sought to identify disease-stage specific biomarkers.
Methods: Using TaqMan microfluidics arrays and a statistical consensus approach, we assessed plasma levels of 185 neurodegeneration-related microRNA, in cohorts of cognitively normal amyloid β-positive (CN-Aβ+), mild cognitive impairment (MCI), and Alzheimer's disease (AD) participants, relative to their respective controls.
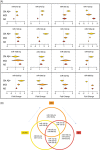
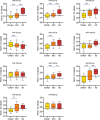
Results: Distinct disease stage microRNA biomarkers were identified, shown to predict membership of the groups (area under the curve [AUC] >0.8) and were altered dynamically with AD progression in a longitudinal study. Bioinformatics demonstrated that these microRNA target known AD-related pathways, such as the Phosphoinositide 3-kinase (PI3K-Akt) signalling pathway. Furthermore, a significant correlation was found between miR-27a-3p, miR-27b-3p, and miR-324-5p and amyloid beta load.
Discussion: Our results show that microRNA signatures alter throughout the progression of AD, reflect the underlying disease pathology, and may prove to be useful diagnostic markers.
Keywords: Alzheimer's disease; biomarker; disease progression; early diagnostic; microRNA; plasma.
© 2021 The Authors. Alzheimer's & Dementia: Diagnosis, Assessment & Disease Monitoring published by Wiley Periodicals, LLC on behalf of Alzheimer's Association.
Conflict of interest statement
Helen Tsui: nothing to disclose Chris J. Fowler: nothing to disclose Colin L. Masters: nothing to disclose Ralph N. Martins: nothing to disclose Nicholas J. Cutfield: Attended conferences and workshops for which costs were subsidized, but received no direct payments. Allergan‐sponsored Movement disorders injectors workshop, Hamilton NZ (2019); Biogen and Roche‐sponsored Multiple Sclerosis meetings (2018, 2019, 2020), (2019); Biogen‐sponsored Multiple Sclerosis ATLAS meeting, Sydney Australia; Sanofi American Academy of Neurology meeting, California, USA.
Figures


References
-
- Moreno‐Grau S, de Rojas I, Hernández I, et al. Genome‐wide association analysis of dementia and its clinical endophenotypes reveal novel loci associated with Alzheimer's disease and three causality networks: the GR@ACE project. Alzheimer's & Dement: J Alzheimer's Assoc. 2019;15(10):1333‐1347. - PubMed
-
- Blennow K, Mattsson N, Schöll M, Hansson O, Zetterberg H. Amyloid biomarkers in Alzheimer's disease. Trends in Pharmacol Sci. 2015;36(5):297‐309. - PubMed
-
- Chételat G, Villemagne VL, Villain N, et al. Accelerated cortical atrophy in cognitively normal elderly with high β‐amyloid deposition. Neurol. 2012;78(7):477‐484. - PubMed
-
- Nakamura A, Kaneko N, Villemagne VL, et al. High performance plasma amyloid‐β biomarkers for Alzheimer's disease. Nature. 2018;554(7691):249‐254. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
