TORC1 inactivation promotes APC/C-dependent mitotic slippage in yeast and human cells
- PMID: 35141499
- PMCID: PMC8814761
- DOI: 10.1016/j.isci.2021.103675
TORC1 inactivation promotes APC/C-dependent mitotic slippage in yeast and human cells
Abstract
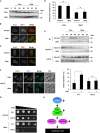
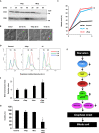
Unsatisfied kinetochore-microtubule attachment activates the spindle assembly checkpoint to inhibit the metaphase-anaphase transition. However, some cells eventually override mitotic arrest by mitotic slippage. Here, we show that inactivation of TORC1 kinase elicits mitotic slippage in budding yeast and human cells. Yeast mitotic slippage was accompanied with aberrant aspects, such as degradation of the nucleolar protein Net1, release of phosphatase Cdc14, and anaphase-promoting complex/cyclosome (APC/C)-Cdh1-dependent degradation of securin and cyclin B in metaphase. This mitotic slippage caused chromosome instability. In human cells, mammalian TORC1 (mTORC1) inactivation also invoked mitotic slippage, indicating that TORC1 inactivation-induced mitotic slippage is conserved from yeast to mammalian cells. However, the invoked mitotic slippage in human cells was not dependent on APC/C-Cdh1. This study revealed an unexpected involvement of TORC1 in mitosis and provides information on undesirable side effects of the use of TORC1 inhibitors as immunosuppressants and anti-tumor drugs.
Keywords: Biological sciences; Cell biology; Molecular biology.
© 2021.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Astrinidis A., Senapedis W., Henske E.P. Hamartin, the tuberous sclerosis complex 1 gene product, interacts with polo-like kinase 1 in a phosphorylation-dependent manner. Hum. Mol. Genet. 2006;15:287–297. - PubMed
-
- Azzam R., Chen S.L., Shou W., Mah A.S., Alexandru G., Nasmyth K., Annan R.S., Carr S.A., Deshaies R.J. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science. 2004;305:516–519. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

