Upregulation of CD14 in mesenchymal stromal cells accelerates lipopolysaccharide-induced response and enhances antibacterial properties
- PMID: 35141503
- PMCID: PMC8814754
- DOI: 10.1016/j.isci.2022.103759
Upregulation of CD14 in mesenchymal stromal cells accelerates lipopolysaccharide-induced response and enhances antibacterial properties
Abstract
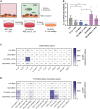
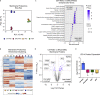
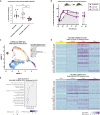
Mesenchymal stromal cells (MSCs) have broad-ranging therapeutic properties, including the ability to inhibit bacterial growth and resolve infection. However, the genetic mechanisms regulating these antibacterial properties in MSCs are largely unknown. Here, we utilized a systems-based approach to compare MSCs from different genetic backgrounds that displayed differences in antibacterial activity. Although both MSCs satisfied traditional MSC-defining criteria, comparative transcriptomics and quantitative membrane proteomics revealed two unique molecular profiles. The antibacterial MSCs responded rapidly to bacterial lipopolysaccharide (LPS) and had elevated levels of the LPS co-receptor CD14. CRISPR-mediated overexpression of endogenous CD14 in MSCs resulted in faster LPS response and enhanced antibacterial activity. Single-cell RNA sequencing of CD14-upregulated MSCs revealed a shift in transcriptional ground state and a more uniform LPS-induced response. Our results highlight the impact of genetic background on MSC phenotypic diversity and demonstrate that overexpression of CD14 can prime these cells to be more responsive to bacterial challenge.
Keywords: Cell biology; Molecular biology; Omics.
© 2022 The Authors.
Conflict of interest statement
The authors do not have any competing interests to report.
Figures



Similar articles
-
lncRNA-mRNA expression profiles and functional networks of mesenchymal stromal cells involved in monocyte regulation.Stem Cell Res Ther. 2019 Jul 16;10(1):207. doi: 10.1186/s13287-019-1306-x. Stem Cell Res Ther. 2019. PMID: 31311593 Free PMC article.
-
Prolonged exposure to bacterial toxins downregulated expression of toll-like receptors in mesenchymal stromal cell-derived osteoprogenitors.BMC Cell Biol. 2008 Sep 18;9:52. doi: 10.1186/1471-2121-9-52. BMC Cell Biol. 2008. PMID: 18799018 Free PMC article.
-
Comparison of Antibacterial and Immunological Properties of Mesenchymal Stem/Stromal Cells from Equine Bone Marrow, Endometrium, and Adipose Tissue.Stem Cells Dev. 2018 Nov 1;27(21):1518-1525. doi: 10.1089/scd.2017.0241. Epub 2018 Sep 6. Stem Cells Dev. 2018. PMID: 30044182 Free PMC article.
-
Human mesenchymal stromal cells express CD14 cross-reactive epitopes.Cytometry A. 2011 Aug;79(8):635-45. doi: 10.1002/cyto.a.21073. Epub 2011 Jul 6. Cytometry A. 2011. PMID: 21735544
-
Mitochondrial Transfer and Regulators of Mesenchymal Stromal Cell Function and Therapeutic Efficacy.Front Cell Dev Biol. 2020 Dec 7;8:603292. doi: 10.3389/fcell.2020.603292. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33365311 Free PMC article. Review.
Cited by
-
Amnion responses to intrauterine inflammation and effects of inhibition of TNF signaling in preterm Rhesus macaque.iScience. 2023 Oct 6;26(11):108118. doi: 10.1016/j.isci.2023.108118. eCollection 2023 Nov 17. iScience. 2023. PMID: 37953944 Free PMC article.
-
Immune Activated Cellular Therapy for Drug Resistant Infections: Rationale, Mechanisms, and Implications for Veterinary Medicine.Vet Sci. 2022 Nov 4;9(11):610. doi: 10.3390/vetsci9110610. Vet Sci. 2022. PMID: 36356087 Free PMC article. Review.
-
Ibrutinib Prevents Acute Lung Injury via Multi-Targeting BTK, FLT3 and EGFR in Mice.Int J Mol Sci. 2022 Nov 3;23(21):13478. doi: 10.3390/ijms232113478. Int J Mol Sci. 2022. PMID: 36362264 Free PMC article.
References
-
- Ackerman W.E.T., Summerfield T.L., Vandre D.D., Robinson J.M., Kniss D.A. Nuclear factor-kappa B regulates inducible prostaglandin E synthase expression in human amnion mesenchymal cells. Biol. Reprod. 2008;78:68–76. - PubMed
-
- Bahrami S., Drablos F. Gene regulation in the immediate-early response process. Adv. Biol. Regul. 2016;62:37–49. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

