An ensemble approach to the structure-function problem in microbial communities
- PMID: 35141504
- PMCID: PMC8810406
- DOI: 10.1016/j.isci.2022.103761
An ensemble approach to the structure-function problem in microbial communities
Abstract
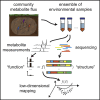
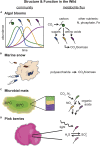
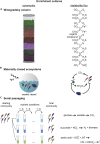
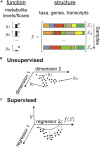
The metabolic activity of microbial communities plays a primary role in the flow of essential nutrients throughout the biosphere. Molecular genetics has revealed the metabolic pathways that model organisms utilize to generate energy and biomass, but we understand little about how the metabolism of diverse, natural communities emerges from the collective action of its constituents. We propose that quantifying and mapping metabolic fluxes to sequencing measurements of genomic, taxonomic, or transcriptional variation across an ensemble of diverse communities, either in the laboratory or in the wild, can reveal low-dimensional descriptions of community structure that can explain or predict their emergent metabolic activity. We survey the types of communities for which this approach might be best suited, review the analytical techniques available for quantifying metabolite fluxes in communities, and discuss what types of data analysis approaches might be lucrative for learning the structure-function mapping in communities from these data.
Keywords: Biophysics; Microbial metabolism; Microbiology.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Aitchison J. The statistical analysis of compositional data. J. R. Stat. Soc. Ser. B (Methodological) 1982;44:139–160.
-
- Aitchison J., Barceló-Vidal C., Martín-Fernández J.A., Pawlowsky-Glahn V. Logratio analysis and compositional distance. Math. Geol. 2000;32:271–275.
-
- Alldredge A.L., Silver M.W. Characteristics, dynamics and significance of marine snow. Prog. Oceanogr. 1988;20:41–82.
Publication types
LinkOut - more resources
Full Text Sources

