Inducers of the NF-κB pathways impair hepatitis delta virus replication and strongly decrease progeny infectivity in vitro
- PMID: 35141510
- PMCID: PMC8792426
- DOI: 10.1016/j.jhepr.2021.100415
Inducers of the NF-κB pathways impair hepatitis delta virus replication and strongly decrease progeny infectivity in vitro
Abstract
Background & aims: HDV superinfection of chronically HBV-infected patients is the most aggressive form of chronic viral hepatitis, with an accelerated progression towards fibrosis/cirrhosis and increased risk of liver failure, hepatocellular carcinoma, and death. While HDV infection is not susceptible to available direct anti-HBV drugs, suboptimal responses are obtained with interferon-α-based therapies, and the number of investigational drugs remains limited. We therefore analyzed the effect of several innate immune stimulators on HDV replication in infected hepatocytes.
Methods: We used in vitro models of HDV and HBV infection based on primary human hepatocytes (PHHs) and the non-transformed HepaRG cell line that are relevant to explore new innate immune therapies.
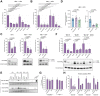
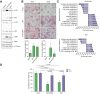
Results: We describe here, for the first time, anti-HDV effects of Pam3CSK4 and BS1, agonists of Toll-like receptor (TLR)-1/2, and the lymphotoxin-β receptor (LTβR), respectively. Both types of agonists induced dose-dependent reductions of total intracellular HDV genome and antigenome RNA and of HDV protein levels, without toxicity in cells monoinfected with HDV or co/superinfected with HBV. Moreover, both molecules negatively affected HDV progeny release and strongly decreased their specific infectivity. The latter effect is particularly important since HDV is thought to persist in humans through constant propagation.
Conclusions: Immune-modulators inducing NF-κB pathways in hepatocytes can inhibit HDV replication and should be further evaluated as a possible therapeutic approach in chronically HBV/HDV-infected patients.
Lay summary: Hepatitis delta virus causes the most severe form of viral hepatitis. Despite positive recent developments, effective treatments remain a major clinical need. Herein, we show that immune-modulators that trigger the NF-κB pathways could be effective for the treatment of hepatitis delta infections.
Keywords: HDV-AG(s), HDV anti-genome(s); HDV-G(s), HDV genome(s); Hepatitis B virus; Hepatitis D virus; IFN, interferon; IL-, interleukin-; L-HDAg, large HDV antigen; LTβR, lymphotoxin-β receptor; NF-κB; NTCP, Na+-taurocholate cotransporting polypeptide; PHH, primary human hepatocyte; Peg-IFN-α, pegylated interferon-α; RNP, ribonucleoprotein; S-HDAg, small HDV antigen; TLR, Toll-like receptor; TNF, tumor necrosis factor; antiviral activity; dHepaRG, differentiated HepaRG cells; hepatocytes; lymphotoxin beta receptor; rh, recombinant human; toll-like receptor; vge, viral genome equivalent.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest that pertain to this work. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures

Similar articles
-
Strain-specific responsiveness of hepatitis D virus to interferon-alpha treatment.JHEP Rep. 2023 Jan 24;5(4):100673. doi: 10.1016/j.jhepr.2023.100673. eCollection 2023 Apr. JHEP Rep. 2023. PMID: 36908749 Free PMC article.
-
HDV RNA replication is associated with HBV repression and interferon-stimulated genes induction in super-infected hepatocytes.Antiviral Res. 2016 Dec;136:19-31. doi: 10.1016/j.antiviral.2016.10.006. Epub 2016 Oct 19. Antiviral Res. 2016. PMID: 27771387
-
Farnesoid X receptor alpha ligands inhibit HDV in vitro replication and virion infectivity.Hepatol Commun. 2023 Apr 14;7(5):e0078. doi: 10.1097/HC9.0000000000000078. eCollection 2023 May 1. Hepatol Commun. 2023. PMID: 37058078 Free PMC article.
-
Future treatments for hepatitis delta virus infection.Liver Int. 2020 Feb;40 Suppl 1:54-60. doi: 10.1111/liv.14356. Liver Int. 2020. PMID: 32077603 Review.
-
Innate immune recognition and modulation in hepatitis D virus infection.World J Gastroenterol. 2020 Jun 7;26(21):2781-2791. doi: 10.3748/wjg.v26.i21.2781. World J Gastroenterol. 2020. PMID: 32550754 Free PMC article. Review.
Cited by
-
Host Immune Response in Chronic Hepatitis Delta: Implications for Pathogenesis and Therapy.Pathogens. 2025 Aug 21;14(8):828. doi: 10.3390/pathogens14080828. Pathogens. 2025. PMID: 40872338 Free PMC article. Review.
-
Chinese medicine in the treatment of chronic hepatitis B: The mechanisms of signal pathway regulation.Heliyon. 2024 Oct 12;10(20):e39176. doi: 10.1016/j.heliyon.2024.e39176. eCollection 2024 Oct 30. Heliyon. 2024. PMID: 39640799 Free PMC article. Review.
-
The dual-specificity kinase DYRK1A interacts with the Hepatitis B virus genome and regulates the production of viral RNA.PLoS One. 2024 Oct 15;19(10):e0311655. doi: 10.1371/journal.pone.0311655. eCollection 2024. PLoS One. 2024. PMID: 39405283 Free PMC article.
-
Toll-Like Receptors in the Immunotherapy Era: Dual-Edged Swords of Tumor Immunity and Clinical Translation.MedComm (2020). 2025 Jul 27;6(8):e70308. doi: 10.1002/mco2.70308. eCollection 2025 Aug. MedComm (2020). 2025. PMID: 40727252 Free PMC article. Review.
-
A novel in vitro system for simultaneous infections with hepatitis B, C, D and E viruses.JHEP Rep. 2025 Feb 28;7(5):101383. doi: 10.1016/j.jhepr.2025.101383. eCollection 2025 May. JHEP Rep. 2025. PMID: 40242313 Free PMC article.
References
-
- Smedile A., Farci P., Verme G., Caredda F., Cargnel A., Caporaso N., et al. Influence of delta infection on severity of hepatitis B. Lancet. 1982;2:945–947. - PubMed
-
- Govindarajan S., Chin K.P., Redeker A.G., Peters R.L. Fulminant B viral hepatitis: role of delta agent. Gastroenterology. 1984;86:1417–1420. - PubMed
-
- Buti M., Homs M., Rodriguez-Frias F., Funalleras G., Jardi R., Sauleda S., et al. Clinical outcome of acute and chronic hepatitis delta over time: a long-term follow-up study. J Viral Hepat. 2011;18:434–442. - PubMed
-
- Lucifora J., Delphin M. Current knowledge on hepatitis delta virus replication. Antivir Res. 2020;179:104812. - PubMed
-
- Yurdaydin C. Treatment of chronic delta hepatitis. Semin Liver Dis. 2012;32:237–244. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

