New developments in the biology of fibroblast growth factors
- PMID: 35142107
- PMCID: PMC10115509
- DOI: 10.1002/wsbm.1549
New developments in the biology of fibroblast growth factors
Abstract
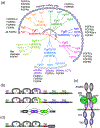
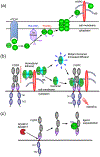
The fibroblast growth factor (FGF) family is composed of 18 secreted signaling proteins consisting of canonical FGFs and endocrine FGFs that activate four receptor tyrosine kinases (FGFRs 1-4) and four intracellular proteins (intracellular FGFs or iFGFs) that primarily function to regulate the activity of voltage-gated sodium channels and other molecules. The canonical FGFs, endocrine FGFs, and iFGFs have been reviewed extensively by us and others. In this review, we briefly summarize past reviews and then focus on new developments in the FGF field since our last review in 2015. Some of the highlights in the past 6 years include the use of optogenetic tools, viral vectors, and inducible transgenes to experimentally modulate FGF signaling, the clinical use of small molecule FGFR inhibitors, an expanded understanding of endocrine FGF signaling, functions for FGF signaling in stem cell pluripotency and differentiation, roles for FGF signaling in tissue homeostasis and regeneration, a continuing elaboration of mechanisms of FGF signaling in development, and an expanding appreciation of roles for FGF signaling in neuropsychiatric diseases. This article is categorized under: Cardiovascular Diseases > Molecular and Cellular Physiology Neurological Diseases > Molecular and Cellular Physiology Congenital Diseases > Stem Cells and Development Cancer > Stem Cells and Development.
Keywords: fibroblast growth factors; organogenesis; receptor tyrosine kinase; regeneration; tyrosine kinase inhibitors.
© 2022 Wiley Periodicals LLC.
Conflict of interest statement
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
Figures

Similar articles
-
Targeting Drugs Against Fibroblast Growth Factor(s)-Induced Cell Signaling.Curr Drug Targets. 2021;22(2):214-240. doi: 10.2174/1389450121999201012201926. Curr Drug Targets. 2021. PMID: 33045958 Review.
-
Non-canonical fibroblast growth factor signalling in angiogenesis.Cardiovasc Res. 2008 May 1;78(2):223-31. doi: 10.1093/cvr/cvm086. Epub 2007 Dec 4. Cardiovasc Res. 2008. PMID: 18056763 Review.
-
Small molecule inhibition of fibroblast growth factor receptors in cancer.Cytokine Growth Factor Rev. 2013 Oct;24(5):467-75. doi: 10.1016/j.cytogfr.2013.05.002. Epub 2013 Jul 2. Cytokine Growth Factor Rev. 2013. PMID: 23830577 Review.
-
The Role of Fibroblast Growth Factor (FGF) Signaling in Tissue Repair and Regeneration.Cells. 2021 Nov 19;10(11):3242. doi: 10.3390/cells10113242. Cells. 2021. PMID: 34831463 Free PMC article. Review.
-
The Fibroblast Growth Factor signaling pathway.Wiley Interdiscip Rev Dev Biol. 2015 May-Jun;4(3):215-66. doi: 10.1002/wdev.176. Epub 2015 Mar 13. Wiley Interdiscip Rev Dev Biol. 2015. PMID: 25772309 Free PMC article. Review.
Cited by
-
Fibroblast growth factor-9 expression in airway epithelial cells amplifies the type I interferon response and alters influenza A virus pathogenesis.PLoS Pathog. 2022 Jun 8;18(6):e1010228. doi: 10.1371/journal.ppat.1010228. eCollection 2022 Jun. PLoS Pathog. 2022. PMID: 35675358 Free PMC article.
-
Cordycepin Activates Autophagy to Suppress FGF9-induced TM3 Mouse Leydig Progenitor Cell Proliferation.Cancer Genomics Proteomics. 2024 Nov-Dec;21(6):630-644. doi: 10.21873/cgp.20479. Cancer Genomics Proteomics. 2024. PMID: 39467624 Free PMC article.
-
Endogenous FGF1 Deficiency Aggravates Doxorubicin-Induced Hepatotoxicity.Toxics. 2023 Nov 12;11(11):925. doi: 10.3390/toxics11110925. Toxics. 2023. PMID: 37999577 Free PMC article.
-
Computer-assisted stabilization of fibroblast growth factor FGF-18.Comput Struct Biotechnol J. 2023 Oct 9;21:5144-5152. doi: 10.1016/j.csbj.2023.10.009. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37920818 Free PMC article.
-
Less, but More: New Insights From Appendicularians on Chordate Fgf Evolution and the Divergence of Tunicate Lifestyles.Mol Biol Evol. 2025 Jan 6;42(1):msae260. doi: 10.1093/molbev/msae260. Mol Biol Evol. 2025. PMID: 39686543 Free PMC article.
References
-
- Abu-Issa R, Smyth G, Smoak I, Yamamura K, & Meyers EN (2002). Fgf8 is required for pharyngeal arch and cardiovascular development in the mouse. Development, 129(19), 4613–4625. - PubMed
-
- Abu-Odeh M, Zhang Y, Reilly SM, Ebadat N, Keinan O, Valentine JM, Hafezi-Bakhtiari M, Ashayer H, Mamoun L, Zhou X, Zhang J, Yu RT, Dai Y, Liddle C, Downes M, Evans RM, Kliewer SA, Mangelsdorf DJ, & Saltiel AR (2021). FGF21 promotes thermogenic gene expression as an autocrine factor in adipocytes. Cell Reports, 35(13), 109331. 10.1016/j.celrep.2021.109331 - DOI - PMC - PubMed
-
- Achilleos A, Huffman NT, Marcinkiewicyz E, Seidah NG, Chen Q, Dallas SL, Trainor PA, & Gorski JP (2015). MBTPS1/SKI-1/S1P proprotein convertase is required for ECM signaling and axial elongation during somitogenesis and vertebral developmentdagger. Human Molecular Genetics, 24(10), 2884–2898. 10.1093/hmg/ddv050 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

