From environmental adaptation to host survival: Attributes that mediate pathogenicity of Candida auris
- PMID: 35142597
- PMCID: PMC8837256
- DOI: 10.1080/21505594.2022.2026037
From environmental adaptation to host survival: Attributes that mediate pathogenicity of Candida auris
Abstract
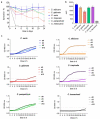
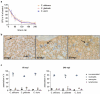
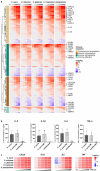
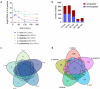
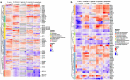
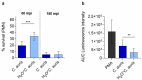
Candida species are a major cause of invasive fungal infections. While Candida albicans, C. glabrata, C. parapsilosis, and C. tropicalis are the most dominant species causing life-threatening candidiasis, C. auris recently emerged as a new species causing invasive infections with high rates of clinical treatment failures. To mimic initial phases of systemic Candida infections with dissemination via the bloodstream and to elucidate the pathogenic potential of C. auris, we used an ex vivo whole blood infection model. Similar to other clinically relevant Candida spp., C. auris is efficiently killed in human blood, but showed characteristic patterns of immune cell association, survival rates, and cytokine induction. Dual-species transcriptional profiling of C. auris-infected blood revealed a unique C. auris gene expression program during infection, while the host response proofed similar and conserved compared to other Candida species. C. auris-specific responses included adaptation and survival strategies, such as counteracting oxidative burst of immune cells, but also expression of potential virulence factors, (drug) transporters, and cell surface-associated genes. Despite comparable pathogenicity to other Candida species in our model, C. auris-specific transcriptional adaptations as well as its increased stress resistance and long-term environmental survival, likely contribute to the high risk of contamination and distribution in a nosocomial setting. Moreover, infections of neutrophils with pre-starved C. auris cells suggest that environmental preconditioning can have modulatory effects on the early host interaction. In summary, we present novel insights into C. auris pathogenicity, revealing adaptations to human blood and environmental niches distinctive from other Candida species.
Keywords: Candida auris; ROS; blood infection; cell surface; neutrophils; transcriptional profiling; transporter; virulence factor.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



References
-
- Perlroth J, Choi B, Spellberg B.. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol. 2007;45(4):321–346. - PubMed
-
- Wisplinghoff H, Ebbers J, Geurtz L, et al. Nosocomial bloodstream infections due to Candida spp. in the USA: species distribution, clinical features and antifungal susceptibilities. Int J Antimicrob Agents. 2014;43(1):78–81. - PubMed
-
- Guinea J. Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect. 2014;20(6):5–10. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
