Broad anti-SARS-CoV-2 antibody immunity induced by heterologous ChAdOx1/mRNA-1273 vaccination
- PMID: 35143256
- PMCID: PMC8939765
- DOI: 10.1126/science.abn2688
Broad anti-SARS-CoV-2 antibody immunity induced by heterologous ChAdOx1/mRNA-1273 vaccination
Abstract
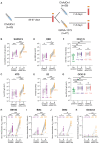
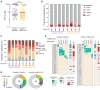
Heterologous prime-boost immunization strategies have the potential to augment COVID-19 vaccine efficacy. We longitudinally profiled severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S)-specific serological and memory B cell (MBC) responses in individuals who received either homologous (ChAdOx1:ChAdOx1) or heterologous (ChAdOx1:mRNA-1273) prime-boost vaccination. Heterologous messenger RNA (mRNA) booster immunization induced higher serum neutralizing antibody and MBC responses against SARS-CoV-2 variants of concern (VOCs) compared with that of homologous ChAdOx1 boosting. Specificity mapping of circulating B cells revealed that mRNA-1273 boost immunofocused ChAdOx1-primed responses onto epitopes expressed on prefusion-stabilized S. Monoclonal antibodies isolated from mRNA-1273-boosted participants displayed overall higher binding affinities and increased breadth of reactivity against VOCs relative to those isolated from ChAdOx1-boosted individuals. Overall, the results provide molecular insight into the enhanced quality of the B cell response induced after heterologous mRNA booster vaccination.
Figures


Similar articles
-
Follow-Up and Comparative Assessment of SARS-CoV-2 IgA, IgG, Neutralizing, and Total Antibody Responses After BNT162b2 or mRNA-1273 Heterologous Booster Vaccination.Influenza Other Respir Viruses. 2024 May;18(5):e13290. doi: 10.1111/irv.13290. Influenza Other Respir Viruses. 2024. PMID: 38706402 Free PMC article.
-
Kinetics of the Antibody Response to Boostering With Three Different Vaccines Against SARS-CoV-2.Front Immunol. 2022 Jan 19;13:811020. doi: 10.3389/fimmu.2022.811020. eCollection 2022. Front Immunol. 2022. PMID: 35126395 Free PMC article.
-
Heterologous ChAdOx1 nCoV-19 and BNT162b2 prime-boost vaccination elicits potent neutralizing antibody responses and T cell reactivity against prevalent SARS-CoV-2 variants.EBioMedicine. 2022 Jan;75:103761. doi: 10.1016/j.ebiom.2021.103761. Epub 2021 Dec 17. EBioMedicine. 2022. PMID: 34929493 Free PMC article. Clinical Trial.
-
The Effects of Heterologous Immunization with Prime-Boost COVID-19 Vaccination against SARS-CoV-2.Vaccines (Basel). 2021 Oct 11;9(10):1163. doi: 10.3390/vaccines9101163. Vaccines (Basel). 2021. PMID: 34696271 Free PMC article. Review.
-
To mix or not to mix? A rapid systematic review of heterologous prime-boost covid-19 vaccination.Expert Rev Vaccines. 2021 Oct;20(10):1211-1220. doi: 10.1080/14760584.2021.1971522. Epub 2021 Sep 1. Expert Rev Vaccines. 2021. PMID: 34415818 Free PMC article.
Cited by
-
Omicron-specific mRNA vaccine induced cross-protective immunity against ancestral SARS-CoV-2 infection with low neutralizing antibodies.J Med Virol. 2023 Jan;95(1):e28370. doi: 10.1002/jmv.28370. J Med Virol. 2023. PMID: 36458553 Free PMC article.
-
Three immunizations with Novavax's protein vaccines increase antibody breadth and provide durable protection from SARS-CoV-2.NPJ Vaccines. 2024 Jan 20;9(1):17. doi: 10.1038/s41541-024-00806-2. NPJ Vaccines. 2024. PMID: 38245545 Free PMC article.
-
Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.Nat Commun. 2023 Jan 19;14(1):311. doi: 10.1038/s41467-023-35949-8. Nat Commun. 2023. PMID: 36658148 Free PMC article.
-
Enhancing Anti-SARS-CoV-2 Neutralizing Immunity by Genetic Delivery of Enveloped Virus-like Particles Displaying SARS-CoV-2 Spikes.Vaccines (Basel). 2023 Aug 31;11(9):1438. doi: 10.3390/vaccines11091438. Vaccines (Basel). 2023. PMID: 37766115 Free PMC article.
-
Evidence for the heterologous benefits of prior BCG vaccination on COVISHIELD™ vaccine-induced immune responses in SARS-CoV-2 seronegative young Indian adults.Front Immunol. 2022 Oct 4;13:985938. doi: 10.3389/fimmu.2022.985938. eCollection 2022. Front Immunol. 2022. PMID: 36268023 Free PMC article.
References
-
- Madhi S. A., Baillie V., Cutland C. L., Voysey M., Koen A. L., Fairlie L., Padayachee S. D., Dheda K., Barnabas S. L., Bhorat Q. E., Briner C., Kwatra G., Ahmed K., Aley P., Bhikha S., Bhiman J. N., Bhorat A. E., du Plessis J., Esmail A., Groenewald M., Horne E., Hwa S.-H., Jose A., Lambe T., Laubscher M., Malahleha M., Masenya M., Masilela M., McKenzie S., Molapo K., Moultrie A., Oelofse S., Patel F., Pillay S., Rhead S., Rodel H., Rossouw L., Taoushanis C., Tegally H., Thombrayil A., van Eck S., Wibmer C. K., Durham N. M., Kelly E. J., Villafana T. L., Gilbert S., Pollard A. J., de Oliveira T., Moore P. L., Sigal A., Izu A.; NGS-SA Group; Wits-VIDA COVID Group , Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. N. Engl. J. Med. 384, 1885–1898 (2021). 10.1056/NEJMoa2102214 - DOI - PMC - PubMed
-
- Hansen C. H., et al. ., Vaccine effectiveness against SARS-CoV-2 infection with the Omicron or Delta variants following a two-dose or booster BNT162b2 or mRNA-1273 vaccination series: A Danish cohort study. medRxiv, 2021.2012.2020.21267966 (2021).10.1101/2021.12.20.21267966 - DOI
-
- Simpson C. R., Shi T., Vasileiou E., Katikireddi S. V., Kerr S., Moore E., McCowan C., Agrawal U., Shah S. A., Ritchie L. D., Murray J., Pan J., Bradley D. T., Stock S. J., Wood R., Chuter A., Beggs J., Stagg H. R., Joy M., Tsang R. S. M., de Lusignan S., Hobbs R., Lyons R. A., Torabi F., Bedston S., O’Leary M., Akbari A., McMenamin J., Robertson C., Sheikh A., First-dose ChAdOx1 and BNT162b2 COVID-19 vaccines and thrombocytopenic, thromboembolic and hemorrhagic events in Scotland. Nat. Med. 27, 1290–1297 (2021). 10.1038/s41591-021-01408-4 - DOI - PMC - PubMed
-
- Schulz J. B., Berlit P., Diener H.-C., Gerloff C., Greinacher A., Klein C., Petzold G. C., Piccininni M., Poli S., Röhrig R., Steinmetz H., Thiele T., Kurth T., German Society of Neurology SARS-CoV-2 Vaccination Study Group , COVID-19 vaccine-associated cerebral venous thrombosis in Germany. Ann. Neurol. 90, 627–639 (2021). 10.1002/ana.26172 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

