The B.1.427/1.429 (epsilon) SARS-CoV-2 variants are more virulent than ancestral B.1 (614G) in Syrian hamsters
- PMID: 35143587
- PMCID: PMC8865701
- DOI: 10.1371/journal.ppat.1009914
The B.1.427/1.429 (epsilon) SARS-CoV-2 variants are more virulent than ancestral B.1 (614G) in Syrian hamsters
Abstract
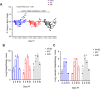
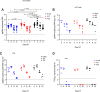
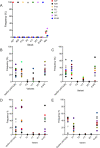
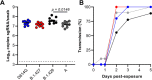
As novel SARS-CoV-2 variants continue to emerge, it is critical that their potential to cause severe disease and evade vaccine-induced immunity is rapidly assessed in humans and studied in animal models. In early January 2021, a novel SARS-CoV-2 variant designated B.1.429 comprising 2 lineages, B.1.427 and B.1.429, was originally detected in California (CA) and it was shown to have enhanced infectivity in vitro and decreased antibody neutralization by plasma from convalescent patients and vaccine recipients. Here we examine the virulence, transmissibility, and susceptibility to pre-existing immunity for B 1.427 and B 1.429 in the Syrian hamster model. We find that both variants exhibit enhanced virulence as measured by increased body weight loss compared to hamsters infected with ancestral B.1 (614G), with B.1.429 causing the most marked body weight loss among the 3 variants. Faster dissemination from airways to parenchyma and more severe lung pathology at both early and late stages were also observed with B.1.429 infections relative to B.1. (614G) and B.1.427 infections. In addition, subgenomic viral RNA (sgRNA) levels were highest in oral swabs of hamsters infected with B.1.429, however sgRNA levels in lungs were similar in all three variants. This demonstrates that B.1.429 replicates to higher levels than ancestral B.1 (614G) or B.1.427 in the oropharynx but not in the lungs. In multi-virus in-vivo competition experiments, we found that B.1. (614G), epsilon (B.1.427/B.1.429) and gamma (P.1) dramatically outcompete alpha (B.1.1.7), beta (B.1.351) and zeta (P.2) in the lungs. In the nasal cavity, B.1. (614G), gamma, and epsilon dominate, but the highly infectious alpha variant also maintains a moderate size niche. We did not observe significant differences in airborne transmission efficiency among the B.1.427, B.1.429 and ancestral B.1 (614G) and WA-1 variants in hamsters. These results demonstrate enhanced virulence and high relative oropharyngeal replication of the epsilon (B.1.427/B.1.429) variant in Syrian hamsters compared to an ancestral B.1 (614G) variant.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Update of
-
The B.1.427/1.429 (epsilon) SARS-CoV-2 variants are more virulent than ancestral B.1 (614G) in Syrian hamsters.bioRxiv [Preprint]. 2021 Aug 25:2021.08.25.457626. doi: 10.1101/2021.08.25.457626. bioRxiv. 2021. Update in: PLoS Pathog. 2022 Feb 10;18(2):e1009914. doi: 10.1371/journal.ppat.1009914. PMID: 34462750 Free PMC article. Updated. Preprint.
Similar articles
-
The B.1.427/1.429 (epsilon) SARS-CoV-2 variants are more virulent than ancestral B.1 (614G) in Syrian hamsters.bioRxiv [Preprint]. 2021 Aug 25:2021.08.25.457626. doi: 10.1101/2021.08.25.457626. bioRxiv. 2021. Update in: PLoS Pathog. 2022 Feb 10;18(2):e1009914. doi: 10.1371/journal.ppat.1009914. PMID: 34462750 Free PMC article. Updated. Preprint.
-
Emerging Variants of SARS-CoV-2 and Novel Therapeutics Against Coronavirus (COVID-19).2023 May 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2023 May 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 34033342 Free Books & Documents.
-
Characterization of a new SARS-CoV-2 variant that emerged in Brazil.Proc Natl Acad Sci U S A. 2021 Jul 6;118(27):e2106535118. doi: 10.1073/pnas.2106535118. Proc Natl Acad Sci U S A. 2021. PMID: 34140350 Free PMC article.
-
Enhanced fitness of SARS-CoV-2 variant of concern Alpha but not Beta.Nature. 2022 Feb;602(7896):307-313. doi: 10.1038/s41586-021-04342-0. Epub 2021 Dec 22. Nature. 2022. PMID: 34937050 Free PMC article.
-
Characterization of the SARS-CoV-2 BA.5.5 and BQ.1.1 Omicron variants in mice and hamsters.J Virol. 2023 Sep 28;97(9):e0062823. doi: 10.1128/jvi.00628-23. Epub 2023 Sep 7. J Virol. 2023. PMID: 37676002 Free PMC article.
Cited by
-
Severe Acute Respiratory Syndrome Coronavirus 2 Vasculopathy in a Syrian Golden Hamster Model.Am J Pathol. 2023 Jun;193(6):690-701. doi: 10.1016/j.ajpath.2023.02.013. Epub 2023 Mar 10. Am J Pathol. 2023. PMID: 36906263 Free PMC article.
-
Mutational dynamics of SARS-CoV-2: Impact on future COVID-19 vaccine strategies.Heliyon. 2024 Apr 25;10(9):e30208. doi: 10.1016/j.heliyon.2024.e30208. eCollection 2024 May 15. Heliyon. 2024. PMID: 38707429 Free PMC article. Review.
-
SARS-CoV-2 pathogenesis.Nat Rev Microbiol. 2022 May;20(5):270-284. doi: 10.1038/s41579-022-00713-0. Epub 2022 Mar 30. Nat Rev Microbiol. 2022. PMID: 35354968 Review.
-
Antibody-mediated immunity to SARS-CoV-2 spike.Adv Immunol. 2022;154:1-69. doi: 10.1016/bs.ai.2022.07.001. Epub 2022 Aug 22. Adv Immunol. 2022. PMID: 36038194 Free PMC article. Review.
-
BA.1, BA.2 and BA.2.75 variants show comparable replication kinetics, reduced impact on epithelial barrier and elicit cross-neutralizing antibodies.PLoS Pathog. 2023 Feb 24;19(2):e1011196. doi: 10.1371/journal.ppat.1011196. eCollection 2023 Feb. PLoS Pathog. 2023. PMID: 36827451 Free PMC article.
References
-
- CDC. SARS-CoV-2 Variant Classifications and Definitions. 2021; Volume. Avalable from: https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

