A protease-activatable luminescent biosensor and reporter cell line for authentic SARS-CoV-2 infection
- PMID: 35143592
- PMCID: PMC8865646
- DOI: 10.1371/journal.ppat.1010265
A protease-activatable luminescent biosensor and reporter cell line for authentic SARS-CoV-2 infection
Abstract
Efforts to define serological correlates of protection against COVID-19 have been hampered by the lack of a simple, scalable, standardised assay for SARS-CoV-2 infection and antibody neutralisation. Plaque assays remain the gold standard, but are impractical for high-throughput screening. In this study, we show that expression of viral proteases may be used to quantitate infected cells. Our assays exploit the cleavage of specific oligopeptide linkers, leading to the activation of cell-based optical biosensors. First, we characterise these biosensors using recombinant SARS-CoV-2 proteases. Next, we confirm their ability to detect viral protease expression during replication of authentic virus. Finally, we generate reporter cells stably expressing an optimised luciferase-based biosensor, enabling viral infection to be measured within 24 h in a 96- or 384-well plate format, including variants of concern. We have therefore developed a luminescent SARS-CoV-2 reporter cell line, and demonstrated its utility for the relative quantitation of infectious virus and titration of neutralising antibodies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
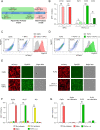
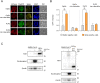
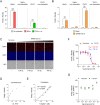
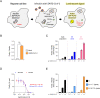
References
-
- Xie X, Muruato AE, Zhang X, Lokugamage KG, Fontes-Garfias CR, Zou J, et al. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat Commun. 2020;11(1):5214. Epub 2020/10/17. doi: 10.1038/s41467-020-19055-7 ; PubMed Central PMCID: PMC7567097. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

