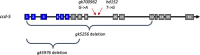
ccd-5, a novel cdk-5 binding partner, regulates pioneer axon guidance in the ventral nerve cord of Caenorhabditis elegans
- PMID: 35143653
- PMCID: PMC8982044
- DOI: 10.1093/genetics/iyac024
ccd-5, a novel cdk-5 binding partner, regulates pioneer axon guidance in the ventral nerve cord of Caenorhabditis elegans
Abstract
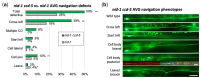
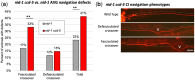
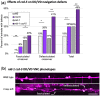
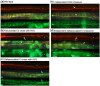
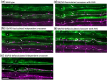
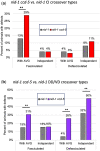
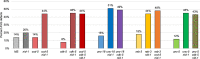
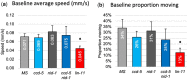
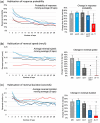
During nervous system development, axons navigate complex environments to reach synaptic targets. Early extending axons must interact with guidance cues in the surrounding tissue, while later extending axons can interact directly with earlier "pioneering" axons, "following" their path. In Caenorhabditis elegans, the AVG neuron pioneers the right axon tract of the ventral nerve cord. We previously found that aex-3, a rab-3 guanine nucleotide exchange factor, is essential for AVG axon navigation in a nid-1 mutant background and that aex-3 might be involved in trafficking of UNC-5, a receptor for the guidance cue UNC-6/netrin. Here, we describe a new gene in this pathway: ccd-5, a putative cdk-5 binding partner. ccd-5 mutants exhibit increased navigation defects of AVG pioneer as well as interneuron and motor neuron follower axons in a nid-1 mutant background. We show that ccd-5 acts in a pathway with cdk-5, aex-3, and unc-5. Navigation defects of follower interneuron and motoneuron axons correlate with AVG pioneer axon defects. This suggests that ccd-5 mostly affects pioneer axon navigation and that follower axon defects are largely a secondary consequence of pioneer navigation defects. To determine the consequences for nervous system function, we assessed various behavioral and movement parameters. ccd-5 single mutants have no significant movement defects, and nid-1 ccd-5 double mutants are less responsive to mechanosensory stimuli compared with nid-1 single mutants. These surprisingly minor defects indicate either a high tolerance for axon guidance defects within the motor circuit and/or an ability to maintain synaptic connections among commonly misguided axons.
Keywords: axon guidance; axon navigation; cdk5; growth cone; nervous system development; pioneer; trafficking.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures




References
-
- Au V, Li-Leger E, Raymant G, Flibotte S, Chen G, Martin K, Fernando L, Doell C, Rosell FI, Wang S, et al.CRISPR/Cas9 methodology for the generation of knockout deletions in Caenorhabditis elegans. G3 (Bethesda). 2019;9(1):135–144. doi:10.1534/g3.118.200778. - DOI - PMC - PubMed
-
- Bhaskar K, Shareef MM, Sharma VM, Shetty APK, Ramamohan Y, Pant HC, Raju TR, Shetty KT.. Co-purification and localization of Munc18-1 (p67) and Cdk5 with neuronal cytoskeletal proteins. Neurochem Int. 2004;44(1):35–44. doi:10.1016/S0197-0186(03)00099-8. - DOI - PubMed
-
- Bhat JM, Hutter H.. Pioneer axon navigation is controlled by AEX-3, a guanine nucleotide exchange factor for RAB-3 in Caenorhabditis elegans. Genetics. 2016;203(3):1235–1247. doi:10.1534/genetics.115.186064. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

