Unveiling the Genetic History of the Maniq, a Primary Hunter-Gatherer Society
- PMID: 35143674
- PMCID: PMC9005329
- DOI: 10.1093/gbe/evac021
Unveiling the Genetic History of the Maniq, a Primary Hunter-Gatherer Society
Abstract
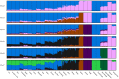
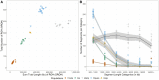
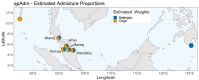
The Maniq of southern Thailand is one of the last remaining practicing hunter-gatherer communities in the world. However, our knowledge on their genetic origins and demographic history is still largely limited. We present here the genotype data covering ∼2.3 million single nucleotide polymorphisms of 11 unrelated Maniq individuals. Our analyses reveal the Maniq to be closely related to the Semang populations of Malaysia (Malay Negritos), who altogether carry an Andamanese-related ancestry linked to the ancient Hòabìnhian hunter-gatherers of Mainland Southeast Asia (MSEA). Moreover, the Maniq possess ∼35% East Asian-related ancestry, likely brought about by recent admixture with surrounding agriculturist communities in the region. In addition, the Maniq exhibit one of the highest levels of genetic differentiation found among living human populations, indicative of their small population size and historical practice of endogamy. Similar to other hunter-gatherer populations of MSEA, we also find the Maniq to possess low levels of Neanderthal ancestry and undetectable levels of Denisovan ancestry. Altogether, we reveal the Maniq to be a Semang group that experienced intense genetic drift and exhibits signs of ancient Hòabìnhian ancestry.
Keywords: Negritos; Semang; Southeast Asia; genetic ancestry; population genetics.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures




References
-
- Barrows DP. 1910. The Negrito and allied types in the Philippines. Am Anthropol. 12(3):358–376.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

