HiCmapTools: a tool to access HiC contact maps
- PMID: 35144531
- PMCID: PMC8832839
- DOI: 10.1186/s12859-022-04589-y
HiCmapTools: a tool to access HiC contact maps
Abstract
Background: With the development of HiC technology, more and more HiC sequencing data have been produced. Although there are dozens of packages that can turn sequencing data into contact maps, there is no appropriate tool to query contact maps in order to extract biological information from HiC datasets.
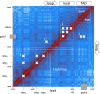
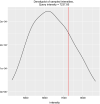
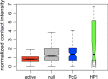
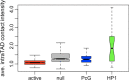
Results: We present HiCmapTools, a tool for biologists to efficiently calculate and analyze HiC maps. The complete program provides multi-query modes and analysis tools. We have validated its utility on two real biological questions: TAD loop and TAD intra-density.
Conclusions: HiCmapTools supports seven access options so that biologists can quantify contact frequency of the interest sites. The tool has been implemented in C++ and R and is freely available at https://github.com/changlabtw/hicmaptools and documented at https://hicmaptools.readthedocs.io/ .
Keywords: 3D genome; Hi-C; Juicer; Topologically Associating Domains (TADs); hicpipe.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

