Whole exome sequencing in Alopecia Areata identifies rare variants in KRT82
- PMID: 35145093
- PMCID: PMC8831607
- DOI: 10.1038/s41467-022-28343-3
Whole exome sequencing in Alopecia Areata identifies rare variants in KRT82
Abstract
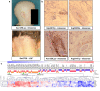
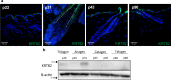
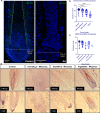
Alopecia areata is a complex genetic disease that results in hair loss due to the autoimmune-mediated attack of the hair follicle. We previously defined a role for both rare and common variants in our earlier GWAS and linkage studies. Here, we identify rare variants contributing to Alopecia Areata using a whole exome sequencing and gene-level burden analyses approach on 849 Alopecia Areata patients compared to 15,640 controls. KRT82 is identified as an Alopecia Areata risk gene with rare damaging variants in 51 heterozygous Alopecia Areata individuals (6.01%), achieving genome-wide significance (p = 2.18E-07). KRT82 encodes a hair-specific type II keratin that is exclusively expressed in the hair shaft cuticle during anagen phase, and its expression is decreased in Alopecia Areata patient skin and hair follicles. Finally, we find that cases with an identified damaging KRT82 variant and reduced KRT82 expression have elevated perifollicular CD8 infiltrates. In this work, we utilize whole exome sequencing to successfully identify a significant Alopecia Areata disease-relevant gene, KRT82, and reveal a proposed mechanism for rare variant predisposition leading to disrupted hair shaft integrity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Islam N, Leung PS, Huntley AC, Gershwin ME. The autoimmune basis of alopecia areata: a comprehensive review. Autoimmun. Rev. 2015;14:81–89. - PubMed
-
- Taheri R, Behnam B, Tousi JA, Azizzade M, Sheikhvatan MR. Triggering role of stressful life events in patients with alopecia areata. Acta Dermatovenerol. Croat. 2012;20:246–250. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

