Copy number amplification of ENSA promotes the progression of triple-negative breast cancer via cholesterol biosynthesis
- PMID: 35145111
- PMCID: PMC8831589
- DOI: 10.1038/s41467-022-28452-z
Copy number amplification of ENSA promotes the progression of triple-negative breast cancer via cholesterol biosynthesis
Abstract
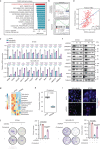
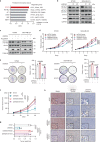
Copy number alterations (CNAs) are pivotal genetic events in triple-negative breast cancer (TNBC). Here, our integrated copy number and transcriptome analysis of 302 TNBC patients reveals that gene alpha-endosulfine (ENSA) exhibits recurrent amplification at the 1q21.3 region and is highly expressed in TNBC. ENSA promotes tumor growth and indicates poor patient survival in TNBC. Mechanistically, we identify ENSA as an essential regulator of cholesterol biosynthesis in TNBC that upregulates the expression of sterol regulatory element-binding transcription factor 2 (SREBP2), a pivotal transcription factor in cholesterol biosynthesis. We confirm that ENSA can increase the level of p-STAT3 (Tyr705) and activated STAT3 binds to the promoter of SREBP2 to promote its transcription. Furthermore, we reveal the efficacy of STAT3 inhibitor Stattic in TNBC with high ENSA expression. In conclusion, the amplification of ENSA at the 1q21.3 region promotes TNBC progression and indicates sensitivity to STAT3 inhibitors.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
PDSS1-Mediated Activation of CAMK2A-STAT3 Signaling Promotes Metastasis in Triple-Negative Breast Cancer.Cancer Res. 2021 Nov 1;81(21):5491-5505. doi: 10.1158/0008-5472.CAN-21-0747. Epub 2021 Aug 18. Cancer Res. 2021. PMID: 34408002
-
PKMYT1 knockdown inhibits cholesterol biosynthesis and promotes the drug sensitivity of triple-negative breast cancer cells to atorvastatin.PeerJ. 2024 Jul 12;12:e17749. doi: 10.7717/peerj.17749. eCollection 2024. PeerJ. 2024. PMID: 39011373 Free PMC article.
-
Copy number amplification of FLAD1 promotes the progression of triple-negative breast cancer through lipid metabolism.Nat Commun. 2025 Feb 1;16(1):1241. doi: 10.1038/s41467-025-56458-w. Nat Commun. 2025. PMID: 39890808 Free PMC article.
-
Epigenetic Reprogramming and Landscape of Transcriptomic Interactions: Impending Therapeutic Interference of Triple-Negative Breast Cancer in Molecular Medicine.Curr Mol Med. 2022;22(10):835-850. doi: 10.2174/1566524021666211206092437. Curr Mol Med. 2022. PMID: 34872474 Review.
-
STAT3 as a potential therapeutic target in triple negative breast cancer: a systematic review.J Exp Clin Cancer Res. 2019 May 14;38(1):195. doi: 10.1186/s13046-019-1206-z. J Exp Clin Cancer Res. 2019. PMID: 31088482 Free PMC article.
Cited by
-
The HOXC10/NOD1/ERK axis drives osteolytic bone metastasis of pan-KRAS-mutant lung cancer.Bone Res. 2024 Aug 27;12(1):47. doi: 10.1038/s41413-024-00350-8. Bone Res. 2024. PMID: 39191757 Free PMC article.
-
Metabolism-regulating non-coding RNAs in breast cancer: roles, mechanisms and clinical applications.J Biomed Sci. 2024 Feb 26;31(1):25. doi: 10.1186/s12929-024-01013-w. J Biomed Sci. 2024. PMID: 38408962 Free PMC article. Review.
-
Cholesterol induces inflammation and reduces glucose utilization.Open Med (Wars). 2023 May 12;18(1):20230701. doi: 10.1515/med-2023-0701. eCollection 2023. Open Med (Wars). 2023. PMID: 37197354 Free PMC article.
-
A phase II study of a doublet metronomic chemotherapy regimen consisting of oral vinorelbine and capecitabine in Chinese women with HER2-negative metastatic breast cancer.Thorac Cancer. 2023 Aug;14(23):2259-2268. doi: 10.1111/1759-7714.15011. Epub 2023 Jul 4. Thorac Cancer. 2023. PMID: 37402471 Free PMC article. Clinical Trial.
-
Ketogenesis promotes triple-negative breast cancer metastasis via calpastatin β-hydroxybutyrylation.Lipids Health Dis. 2024 Nov 12;23(1):371. doi: 10.1186/s12944-024-02364-x. Lipids Health Dis. 2024. PMID: 39533307 Free PMC article.
References
-
- Venkitaraman R. Triple-negative/basal-like breast cancer: clinical, pathologic and molecular features. Expert Rev. Anticancer Ther. 2010;10:199–207. - PubMed
-
- Dent R, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin. Cancer Res. 2007;13:4429–4434. - PubMed
-
- Yin WJ, et al. Clinicopathological features of the triple-negative tumors in Chinese breast cancer patients. Breast Cancer Res Treat. 2009;115:325–333. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

