RNA-seq analysis reveals the genes/pathways responsible for genetic plasticity of rice to varying environmental conditions on direct-sowing and transplanting
- PMID: 35145168
- PMCID: PMC8831524
- DOI: 10.1038/s41598-022-06009-w
RNA-seq analysis reveals the genes/pathways responsible for genetic plasticity of rice to varying environmental conditions on direct-sowing and transplanting
Erratum in
-
Author Correction: RNA-seq analysis reveals the genes/pathways responsible for genetic plasticity of rice to varying environmental conditions on direct-sowing and transplanting.Sci Rep. 2022 Mar 2;12(1):3777. doi: 10.1038/s41598-022-07812-1. Sci Rep. 2022. PMID: 35236908 Free PMC article. No abstract available.
Abstract
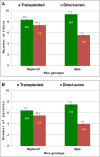
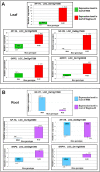
Rice cultivation by transplanting requires plenty of water. It might become a challenging task in future to grow rice by transplanting due to the climatic change, water and labor scarcities. Direct-sown rice (DSR) is emerging as a resource-conserving and climate-smart alternative to transplanted rice (TPR). However, no specific variety has been bred for dry/direct-sown conditions. The present study was undertaken to decipher the molecular basis of genetic plasticity of rice under different planting methods. Comparative RNA-seq analysis revealed a number (6133) of genes exclusively up-regulated in Nagina-22 (N-22) leaf under DSR conditions, compared to that (3538) in IR64 leaf. Several genes up-regulated in N-22 were down-regulated in IR64. Genes for growth-regulation and nutrient-reservoir activities, transcription factors, translational machinery, carbohydrate metabolism, cell cycle/division, and chromatin organization/epigenetic modifications were considerably up-regulated in the leaf of N-22 under DSR conditions. Complementary effects of these factors in rendering genetic plasticity were confirmed by the agronomic/physiological performance of rice cultivar. Thus, growth-regulation/nutrient-reservoir activities, transcription factors, and translational machinery are important molecular factors responsible for the observed genetic plasticity/adaptability of Nagina-22 to different planting methods. This might help to develop molecular markers for DSR breeding, replacing TPR with DSR for better water-productivity, and minimizing greenhouse-gas emission necessary for negative emission agriculture.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







Similar articles
-
Molecular basis of genetic plasticity to varying environmental conditions on growing rice by dry/direct-sowing and exposure to drought stress: Insights for DSR varietal development.Front Plant Sci. 2022 Oct 24;13:1013207. doi: 10.3389/fpls.2022.1013207. eCollection 2022. Front Plant Sci. 2022. PMID: 36352870 Free PMC article.
-
DNA methylome analysis provides insights into gene regulatory mechanism for better performance of rice under fluctuating environmental conditions: epigenomics of adaptive plasticity.Planta. 2023 Nov 22;259(1):4. doi: 10.1007/s00425-023-04272-3. Planta. 2023. PMID: 37993704
-
Agronomic and Environmental Determinants of Direct Seeded Rice in South Asia.Circ Econ Sustain. 2023;3(1):253-290. doi: 10.1007/s43615-022-00173-x. Epub 2022 May 6. Circ Econ Sustain. 2023. PMID: 35573660 Free PMC article. Review.
-
Identification of four functionally important microRNA families with contrasting differential expression profiles between drought-tolerant and susceptible rice leaf at vegetative stage.BMC Genomics. 2015 Sep 15;16(1):692. doi: 10.1186/s12864-015-1851-3. BMC Genomics. 2015. PMID: 26369665 Free PMC article.
-
Current Understanding of Leaf Senescence in Rice.Int J Mol Sci. 2021 Apr 26;22(9):4515. doi: 10.3390/ijms22094515. Int J Mol Sci. 2021. PMID: 33925978 Free PMC article. Review.
Cited by
-
Assessing Loss of Regulatory Divergence, Genome-Transcriptome Incongruence, and Preferential Expression Switching in Abaca × Banana Backcrosses.Genes (Basel). 2022 Aug 6;13(8):1396. doi: 10.3390/genes13081396. Genes (Basel). 2022. PMID: 36011307 Free PMC article.
-
Integrating physiological and multi-omics methods to elucidate heat stress tolerance for sustainable rice production.Physiol Mol Biol Plants. 2024 Jul;30(7):1185-1208. doi: 10.1007/s12298-024-01480-3. Epub 2024 Jul 3. Physiol Mol Biol Plants. 2024. PMID: 39100874 Free PMC article. Review.
-
Molecular basis of genetic plasticity to varying environmental conditions on growing rice by dry/direct-sowing and exposure to drought stress: Insights for DSR varietal development.Front Plant Sci. 2022 Oct 24;13:1013207. doi: 10.3389/fpls.2022.1013207. eCollection 2022. Front Plant Sci. 2022. PMID: 36352870 Free PMC article.
-
The analysis of the genetic loci affecting phenotypic plasticity of soybean isoflavone content by dQTG.seq model.Theor Appl Genet. 2024 Dec 17;138(1):9. doi: 10.1007/s00122-024-04798-4. Theor Appl Genet. 2024. PMID: 39688708
-
Weighted gene co-expression network analysis - based selection of hub genes related to phenolic and volatile compounds and seed coat color in sorghum.BMC Plant Biol. 2025 May 23;25(1):682. doi: 10.1186/s12870-025-06657-w. BMC Plant Biol. 2025. PMID: 40410657 Free PMC article.
References
-
- Seck PA, Diagne A, Mohanty S, Wopereis MC. Crops that feed the world. Rice Food Security. 2012;4:7–24.
-
- Rao AN, Johnson DE, Sivaprasad B, Ladha JK, Mortimer AM. Weed management in direct-seeded rice. Adv. Agron. 2007;93:155–255.
-
- Bouman BAM. How much water does rice use. Rice Today. 2009;69:115–133.
-
- Tuong TP, Bouman BAM, Mortimer M. More rice, less water-integrated approaches for increasing water productivity in irrigated rice-based systems in Asia. Plant Prod. Sci. 2005;8:231–241.
-
- Sun L, et al. Implications of low sowing rate for hybrid rice varieties under dry direct-seeded rice system in Central China. Field Crop Res. 2015;175:87–95.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

