Spatial, temporal and molecular dynamics of swine influenza virus-specific CD8 tissue resident memory T cells
- PMID: 35145208
- PMCID: PMC9038527
- DOI: 10.1038/s41385-021-00478-4
Spatial, temporal and molecular dynamics of swine influenza virus-specific CD8 tissue resident memory T cells
Abstract
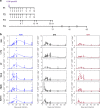
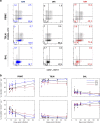
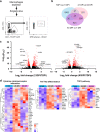
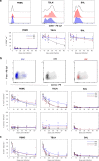
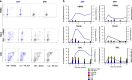
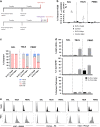
For the first time we have defined naïve, central memory, effector memory and differentiated effector porcine CD8 T cells and analyzed their distribution in lymphoid and respiratory tissues after influenza infection or immunization, using peptide-MHC tetramers of three influenza nucleoprotein (NP) epitopes. The hierarchy of response to the three epitopes changes during the response in different tissues. Most NP-specific CD8 T cells in broncho-alveolar lavage (BAL) and lung are tissue resident memory cells (TRM) that express CD69 and downregulate CD45RA and CCR7. NP-specific cells isolated from BAL express genes characteristic of TRM, but gene expression differs at 7, 21 and 63 days post infection. In all tissues the frequency of NP-specific CD8 cells declines over 63 days almost to background levels but is best maintained in BAL. The kinetic of influenza specific memory CD8 T cell in this natural host species differs from that in small animal models.
© 2022. The Author(s).
Conflict of interest statement
A.T. is named on a patent regarding the use of S-FLU vaccine. The other authors have no financial conflicts of interest.
Figures


References
-
- Harper SA, Fukuda K, Cox NJ, Bridges CB. Advisory Committee on Immunization P. Using live, attenuated influenza vaccine for prevention and control of influenza: supplemental recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm. Rep. 2003;52(RR-13):1–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 220295/Z/20/Z/WT_/Wellcome Trust/United Kingdom
- MR/P021336/1/MRC_/Medical Research Council/United Kingdom
- BB/L001330/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/I/00007036/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/I/0000703/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/I/00007037/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/I/00007039/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/I/00007031/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/I/00007038/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

