Intratumor genetic heterogeneity and clonal evolution to decode endometrial cancer progression
- PMID: 35145232
- PMCID: PMC8956509
- DOI: 10.1038/s41388-022-02221-0
Intratumor genetic heterogeneity and clonal evolution to decode endometrial cancer progression
Abstract
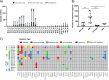
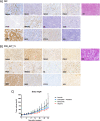
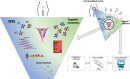
Analyzing different tumor regions by next generation sequencing allows the assessment of intratumor genetic heterogeneity (ITGH), a phenomenon that has been studied widely in some tumor types but has been less well explored in endometrial carcinoma (EC). In this study, we sought to characterize the spatial and temporal heterogeneity of 9 different ECs using whole-exome sequencing, and by performing targeted sequencing validation of the 42 primary tumor regions and 30 metastatic samples analyzed. In addition, copy number alterations of serous carcinomas were assessed by comparative genomic hybridization arrays. From the somatic mutations, identified by whole-exome sequencing, 532 were validated by targeted sequencing. Based on these data, the phylogenetic tree reconstructed for each case allowed us to establish the tumors' evolution and correlate this to tumor progression, prognosis, and the presence of recurrent disease. Moreover, we studied the genetic landscape of an ambiguous EC and the molecular profile obtained was used to guide the selection of a potential personalized therapy for this patient, which was subsequently validated by preclinical testing in patient-derived xenograft models. Overall, our study reveals the impact of analyzing different tumor regions to decipher the ITGH in ECs, which could help make the best treatment decision.
© 2022. The Author(s).
Conflict of interest statement
JSR-F reports receiving personal/consultancy fees from Goldman Sachs, REPARE Therapeutics, Paige.AI and Eli Lilly, membership of the scientific advisory boards of VolitionRx, REPARE Therapeutics and Paige.AI, membership of the Board of Directors of Grupo Oncoclinicas, and ad hoc membership of the scientific advisory boards of Roche Tissue Diagnostics, Ventana Medical Systems, Novartis, Genentech and InVicro, outside the scope of this study. BW reports ad hoc membership of the scientific advisory board of REPARE Therapeutics, outside the scope of this study. The remaining authors have no conflict of interests to declare.
Figures




References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. - PubMed
-
- Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15:10–7. - PubMed
-
- Lax SF, Kurman RJ. A dualistic model for endometrial carcinogenesis based on immunohistochemical and molecular genetic analyses. Verh Dtsch Ges Pathol. 1997;81:228–32. - PubMed
-
- Soslow RA. Endometrial carcinomas with ambiguous features. Semin Diagn Pathol. 2010;27:261–73.. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

