Initial analysis of viral dynamics and circulating viral variants during the mRNA-1273 Phase 3 COVE trial
- PMID: 35145311
- PMCID: PMC9018421
- DOI: 10.1038/s41591-022-01679-5
Initial analysis of viral dynamics and circulating viral variants during the mRNA-1273 Phase 3 COVE trial
Abstract
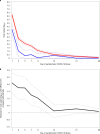
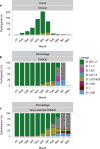
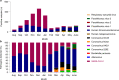
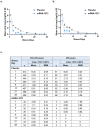
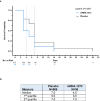
The mRNA-1273 vaccine for coronavirus disease 2019 (COVID-19) demonstrated 93.2% efficacy in reduction of symptomatic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections in the blinded portion of the Phase 3 Coronavirus Efficacy (COVE) trial. While mRNA-1273 demonstrated high efficacy in prevention of COVID-19, including severe disease, its effect on the viral dynamics of SARS-CoV-2 infections is not understood. Here, in exploratory analyses, we assessed the impact of mRNA-1273 vaccination in the ongoing COVE trial (number NCT04470427) on SARS-CoV-2 copy number and shedding, burden of disease and infection, and viral variants. Viral variants were sequenced in all COVID-19 and adjudicated COVID-19 cases (n = 832), from July 2020 in the blinded part A of the study to May 2021 of the open-label part B of the study, in which participants in the placebo arm started to receive the mRNA-1273 vaccine after US Food and Drug Administration emergency use authorization of mRNA-1273 in December 2020. mRNA-1273 vaccination significantly reduced SARS-CoV-2 viral copy number (95% confidence interval) by 100-fold on the day of diagnosis compared with placebo (4.1 (3.4-4.8) versus 6.2 (6.0-6.4) log10 copies per ml). Median times to undetectable viral copies were 4 days for mRNA-1273 and 7 days for placebo. Vaccination also substantially reduced the burden of disease and infection scores. Vaccine efficacies (95% confidence interval) against SARS-CoV-2 variants circulating in the United States during the trial assessed in this post hoc analysis were 82.4% (40.4-94.8%) for variants Epsilon and Gamma and 81.2% (36.1-94.5%) for Epsilon. The detection of other, non-SARS-CoV-2, respiratory viruses during the trial was similar between groups. While additional study is needed, these data show that in SARS-CoV-2-infected individuals, vaccination reduced both the viral copy number and duration of detectable viral RNA, which may be markers for the risk of virus transmission.
© 2022. The Author(s).
Conflict of interest statement
L.R.B. reports grants from NIH/NIAID for the conduct of this study. H.M.E.S. reports grants from NIH during the conduct of the study. I.F. reports receiving research grants from Janssen Pharmaceuticals and Sanofi. E.K. is an employee and founder of Altitude Clinical Consulting and former employee of Crisor LLC, and has consulted, served on advisory boards or received travel reimbursement from Amphastar, AstraZeneca, Boehringer Ingelheim, Chiesi, Connect Biopharma, GlaxoSmithKline, Mylan, Novartis, Sunovion and Theravance. B.E., D.D. and K.M.M. declare no competing interests. R.P., Y.D.P., B.G., G.D., X.Z., B.D., W.D., H.Z. and B.L. report being employees of Moderna, Inc. and may hold stock/stock options in that company. K.K., J.E.T. and F.S. are Moderna consultants.
Figures

Similar articles
-
Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine.N Engl J Med. 2021 Feb 4;384(5):403-416. doi: 10.1056/NEJMoa2035389. Epub 2020 Dec 30. N Engl J Med. 2021. PMID: 33378609 Free PMC article. Clinical Trial.
-
Humoral Immunogenicity of the mRNA-1273 Vaccine in the Phase 3 Coronavirus Efficacy (COVE) Trial.J Infect Dis. 2022 Nov 11;226(10):1731-1742. doi: 10.1093/infdis/jiac188. J Infect Dis. 2022. PMID: 35535503 Free PMC article. Clinical Trial.
-
Long-term safety and effectiveness of mRNA-1273 vaccine in adults: COVE trial open-label and booster phases.Nat Commun. 2024 Aug 29;15(1):7469. doi: 10.1038/s41467-024-50376-z. Nat Commun. 2024. PMID: 39209823 Free PMC article. Clinical Trial.
-
Review of COVID-19 mRNA Vaccines: BNT162b2 and mRNA-1273.J Pharm Pract. 2022 Dec;35(6):947-951. doi: 10.1177/08971900211009650. Epub 2021 Apr 12. J Pharm Pract. 2022. PMID: 33840294 Review.
-
Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: a narrative review.Clin Microbiol Infect. 2022 Feb;28(2):202-221. doi: 10.1016/j.cmi.2021.10.005. Epub 2021 Oct 27. Clin Microbiol Infect. 2022. PMID: 34715347 Free PMC article. Review.
Cited by
-
Evaluation of the analytical performance of the 15-minute point-of-care DASH™ SARS-CoV-2 RT-qPCR test.Diagn Microbiol Infect Dis. 2024 Jan;108(1):116120. doi: 10.1016/j.diagmicrobio.2023.116120. Epub 2023 Oct 21. Diagn Microbiol Infect Dis. 2024. PMID: 37898036 Free PMC article.
-
SARS-CoV-2 Viral Load in the Nasopharynx at Time of First Infection Among Unvaccinated Individuals: A Secondary Cross-Protocol Analysis of 4 Randomized Trials.JAMA Netw Open. 2024 May 1;7(5):e2412835. doi: 10.1001/jamanetworkopen.2024.12835. JAMA Netw Open. 2024. PMID: 38780941 Free PMC article.
-
Immune correlates of protection for SARS-CoV-2, Ebola and Nipah virus infection.Front Immunol. 2023 Apr 17;14:1156758. doi: 10.3389/fimmu.2023.1156758. eCollection 2023. Front Immunol. 2023. PMID: 37153606 Free PMC article. Review.
-
Kinetics of the Antibody Response to Symptomatic SARS-CoV-2 Infection in Vaccinated and Unvaccinated Individuals in the Blinded Phase of the mRNA-1273 COVID-19 Vaccine Efficacy Trial.Open Forum Infect Dis. 2023 Feb 13;10(3):ofad069. doi: 10.1093/ofid/ofad069. eCollection 2023 Mar. Open Forum Infect Dis. 2023. PMID: 36895286 Free PMC article.
-
Efficacy of Messenger RNA-1273 Against Severe Acute Respiratory Syndrome Coronavirus 2 Acquisition in Young Adults From March to December 2021.Open Forum Infect Dis. 2023 Nov 2;10(11):ofad511. doi: 10.1093/ofid/ofad511. eCollection 2023 Nov. Open Forum Infect Dis. 2023. PMID: 38023544 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

