Activation and Immune Regulation Mechanisms of PYHIN Family During Microbial Infection
- PMID: 35145495
- PMCID: PMC8822057
- DOI: 10.3389/fmicb.2021.809412
Activation and Immune Regulation Mechanisms of PYHIN Family During Microbial Infection
Abstract
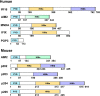
The innate immune system defenses against pathogen infections via patten-recognition receptors (PRRs). PRRs initiate immune responses by recognizing pathogen-associated molecular patterns (PAMPs), including peptidoglycan, lipopolysaccharide, and nucleic acids. Several nucleic acid sensors or families have been identified, such as RIG-I-like receptors (RLRs), Toll-like receptors (TLRs), cyclic GMP-AMP synthase (cGAS), and PYHIN family receptors. In recent years, the PYHIN family cytosolic DNA receptors have increased attention because of their important roles in initiating innate immune responses. The family members in humans include Absent in melanoma 2 (AIM2), IFN-γ inducible protein 16 (IFI16), interferon-inducible protein X (IFIX), and myeloid cell nuclear differentiation antigen (MNDA). The PYHIN family members are also identified in mice, including AIM2, p202, p203, p204, and p205. Herein, we summarize recent advances in understanding the activation and immune regulation mechanisms of the PYHIN family during microbial infection. Furthermore, structural characterizations of AIM2, IFI16, p202, and p204 provide more accurate insights into the signaling mechanisms of PYHIN family receptors. Overall, the molecular details will facilitate the development of reagents to defense against viral infections.
Keywords: AIM2; IFI16; PAMP; PRR; PYHIN family; innate immunity; p202; p204.
Copyright © 2022 Fan, Jiao and Jin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
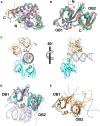

References
Publication types
LinkOut - more resources
Full Text Sources

