Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases
- PMID: 35145997
- PMCID: PMC8822174
- DOI: 10.3389/fmolb.2021.811106
Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases
Abstract
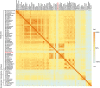
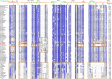
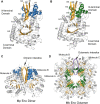
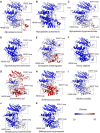
Mycoplasmas are a group of prokaryotes without cell walls that have evolved through several rounds of degenerative evolution. With a low cell DNA G + C content and definitively long genetic lineages, mycoplasmas are thought to be in a state of rapid evolution. However, little associated evidence has been provided. Enolase is a key enzyme in glycolysis that is widely found in all species from the three domains, and it is evolutionarily conserved. In our previous studies, enolase acted as a virulence factor and participated in cell-surface adhesion in Mycoplasma hyopneumoniae. Furthermore, unique loop regions were first found in the crystal structure of Mhp Eno. Here, enolase structures from Mycoplasma pneumoniae and Mycoplasma bovis were determined. An extra helix 7 is specific and conservatively found in almost all mycoplasma enolases, as confirmed by crystal structures and sequence alignment. Particular motifs for helix 7, which is composed of F-K/G-K-L/F-K-X-A-I, have been proposed and could be regarded as molecular markers. To our surprise, the genetic distances between any two mycoplasma enolases were obviously longer than those between the two corresponding species themselves, indicating divergent evolution of mycoplasma enolases, whereas no horizontal gene transfer was detected in mycoplasma enolase genens. Furthermore, different evolutionary patterns were adopted by different loop regions of mycoplasma enolase. Enolases from different Mycoplasma species also showed different affinities for PLG and fibronectin. Our results indicate the rapid and divergent evolution of mycoplasma enolase and mycoplasmas. This study will also aid understanding the independent evolution of Mycoplasma species after separation from their common ancestor.
Keywords: crstal structure; divergent evolution; enolase; mollicutes; mycoplasma.
Copyright © 2022 Chen, Zhao, Gan, Feng, Cui, Xie, Hao, Zhang, Wang, Ran, Wang, Zhang, Li, Zhang, Pang, Xiong and Shao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Featured Species-Specific Loops Are Found in the Crystal Structure of Mhp Eno, a Cell Surface Adhesin From Mycoplasma hyopneumoniae.Front Cell Infect Microbiol. 2019 Jun 13;9:209. doi: 10.3389/fcimb.2019.00209. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31263685 Free PMC article.
-
The surface-localised α-enolase of Mycoplasma suis is an adhesion protein.Vet Microbiol. 2012 Apr 23;156(1-2):88-95. doi: 10.1016/j.vetmic.2011.10.010. Epub 2011 Oct 15. Vet Microbiol. 2012. PMID: 22047714
-
Specific evolution of F1-like ATPases in mycoplasmas.PLoS One. 2012;7(6):e38793. doi: 10.1371/journal.pone.0038793. Epub 2012 Jun 7. PLoS One. 2012. PMID: 22685606 Free PMC article.
-
Horizontal Gene Transfers in Mycoplasmas (Mollicutes).Curr Issues Mol Biol. 2018;29:3-22. doi: 10.21775/cimb.029.003. Epub 2018 Apr 12. Curr Issues Mol Biol. 2018. PMID: 29648541 Review.
-
The central role of lipoproteins in the pathogenesis of mycoplasmoses.Vet Microbiol. 2011 Nov 21;153(1-2):44-50. doi: 10.1016/j.vetmic.2011.05.031. Epub 2011 May 25. Vet Microbiol. 2011. PMID: 21684094 Review.
Cited by
-
Mycoplasmas as Host Pantropic and Specific Pathogens: Clinical Implications, Gene Transfer, Virulence Factors, and Future Perspectives.Front Cell Infect Microbiol. 2022 May 13;12:855731. doi: 10.3389/fcimb.2022.855731. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35646746 Free PMC article. Review.
-
A novel quantitative double antigen sandwich ELISA for detecting total antibodies against Candida albicans enolase 1.Eur J Clin Microbiol Infect Dis. 2024 Sep;43(9):1815-1823. doi: 10.1007/s10096-024-04899-4. Epub 2024 Jul 16. Eur J Clin Microbiol Infect Dis. 2024. PMID: 39012550
-
Transcriptomic Profiling of Fusarium pseudograminearum in Response to Carbendazim, Pyraclostrobin, Tebuconazole, and Phenamacril.J Fungi (Basel). 2023 Mar 8;9(3):334. doi: 10.3390/jof9030334. J Fungi (Basel). 2023. PMID: 36983502 Free PMC article.
-
An alternative vaccine target for bovine Anaplasmosis based on enolase, a moonlighting protein.Front Vet Sci. 2023 Sep 22;10:1225873. doi: 10.3389/fvets.2023.1225873. eCollection 2023. Front Vet Sci. 2023. PMID: 37808115 Free PMC article.
-
KAT2A changes the function of endometrial stromal cells via regulating the succinylation of ENO1.Open Life Sci. 2024 Mar 28;19(1):20220785. doi: 10.1515/biol-2022-0785. eCollection 2024. Open Life Sci. 2024. PMID: 38585644 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

