Hybridization underlies localized trait evolution in cavefish
- PMID: 35146393
- PMCID: PMC8819016
- DOI: 10.1016/j.isci.2022.103778
Hybridization underlies localized trait evolution in cavefish
Abstract
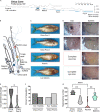
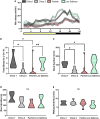
Introgressive hybridization may play an integral role in local adaptation and speciation (Taylor and Larson, 2019). In the Mexican tetra Astyanax mexicanus, cave populations have repeatedly evolved traits including eye loss, sleep loss, and albinism. Of the 30 caves inhabited by A. mexicanus, Chica cave is unique because it contains multiple pools inhabited by putative hybrids between surface and cave populations (Mitchell et al., 1977), providing an opportunity to investigate the impact of hybridization on complex trait evolution. We show that hybridization between cave and surface populations may contribute to localized variation in traits associated with cave evolution, including pigmentation, eye development, and sleep. We also uncover an example of convergent evolution in a circadian clock gene in multiple cavefish lineages and burrowing mammals, suggesting a shared genetic mechanism underlying circadian disruption in subterranean vertebrates. Our results provide insight into the role of hybridization in facilitating phenotypic evolution.
Keywords: Aquatic biology; Aquatic science; Evolutionary biology.
© 2022.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:11.10.1–11.10.33. - PMC - PubMed
-
- Avise J.C., Selander R.K. Evolutionary genetics of cave-dwelling fishes of the genus astyanax. Evolution. 1972;26:1–19. - PubMed
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources

