Protein kinase R is an innate immune sensor of proteotoxic stress via accumulation of cytoplasmic IL-24
- PMID: 35148201
- PMCID: PMC11036408
- DOI: 10.1126/sciimmunol.abi6763
Protein kinase R is an innate immune sensor of proteotoxic stress via accumulation of cytoplasmic IL-24
Abstract
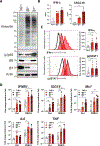
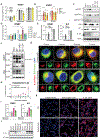
Proteasome dysfunction can lead to autoinflammatory disease associated with elevated type I interferon (IFN-αβ) and NF-κB signaling; however, the innate immune pathway driving this is currently unknown. Here, we identified protein kinase R (PKR) as an innate immune sensor for proteotoxic stress. PKR activation was observed in cellular models of decreased proteasome function and in multiple cell types from patients with proteasome-associated autoinflammatory disease (PRAAS). Furthermore, genetic deletion or small-molecule inhibition of PKR in vitro ameliorated inflammation driven by proteasome deficiency. In vivo, proteasome inhibitor-induced inflammatory gene transcription was blunted in PKR-deficient mice compared with littermate controls. PKR also acted as a rheostat for proteotoxic stress by triggering phosphorylation of eIF2α, which can prevent the translation of new proteins to restore homeostasis. Although traditionally known as a sensor of RNA, under conditions of proteasome dysfunction, PKR sensed the cytoplasmic accumulation of a known interactor, interleukin-24 (IL-24). When misfolded IL-24 egress into the cytosol was blocked by inhibition of the endoplasmic reticulum-associated degradation pathway, PKR activation and subsequent inflammatory signaling were blunted. Cytokines such as IL-24 are normally secreted from cells; therefore, cytoplasmic accumulation of IL-24 represents an internal danger-associated molecular pattern. Thus, we have identified a mechanism by which proteotoxic stress is detected, causing inflammation observed in the disease PRAAS.
Conflict of interest statement
Figures


Comment in
-
Sensing proteotoxic stress.Nat Rev Immunol. 2022 Apr;22(4):205. doi: 10.1038/s41577-022-00702-7. Nat Rev Immunol. 2022. PMID: 35217786 No abstract available.
References
-
- Manthiram K, Zhou Q, Aksentijevich I, Kastner DL, The monogenic autoinflammatory diseases define new pathways in human innate immunity and inflammation. Nat. Immunol 18, 832–842 (2017). - PubMed
-
- Arima K, Kinoshita A, Mishima H, Kanazawa N, Kaneko T, Mizushima T, Ichinose K, Nakamura H, Tsujino A, Kawakami A, Matsunaka M, Kasagi S, Kawano S, Kumagai S, Ohmura K, Mimori T, Hirano M, Ueno S, Tanaka K, Tanaka M, Toyoshima I, Sugino H, Yamakawa A, Tanaka K, Niikawa N, Furukawa F, Murata S, Eguchi K, Ida H, Yoshiura KI, Proteasome assembly defect due to a proteasome subunit β type 8 (PSMB8) mutation causes the autoinflammatory disorder, Nakajo-Nishimura syndrome. Proc. Natl. Acad. Sci. U.S.A 108, 14914–14919 (2011). - PMC - PubMed
-
- Brehm A, Liu Y, Sheikh A, Marrero B, Omoyinmi E, Zhou Q, Montealegre G, Biancotto A, Reinhardt A, Almeida de Jesus A, Pelletier M, Tsai WL, Remmers EF, Kardava L, Hill S, Kim H, Lachmann HJ, Megarbane A, Chae JJ, Brady J, Castillo RD, Brown D, Casano AV, Gao L, Chapelle D, Huang Y, Stone D, Chen Y, Sotzny F, Lee CCR, Kastner DL, Torrelo A, Zlotogorski A, Moir S, Gadina M, McCoy P, Wesley R, Rother K, Hildebrand PW, Brogan P, Krüger E, Aksentijevich I, Goldbach-Mansky R, Additive loss-of-function proteasome subunit mutations in CANDLE/PRAAS patients promote type I IFN production. J. Clin. Invest 125, 4196–4211 (2015). - PMC - PubMed
-
- de Jesus AA, Brehm A, Van Tries R, Pillet P, Parentelli A-S, Sanchez GAM, Deng Z, Paut IK, Goldbach-Mansky R, Krüger E, Novel proteasome assembly chaperone mutations in PSMG2/PAC2 cause the autoinflammatory interferonopathy CANDLE/ PRAAS4. J. Allergy Clin. Immunol 143, 1939–1943.e8 (2019). - PMC - PubMed
-
- Kitamura A, Maekawa Y, Uehara H, Izumi K, Kawachi I, Nishizawa M, Toyoshima Y, Takahashi H, Standley DM, Tanaka K, Hamazaki J, Murata S, Obara K, Toyoshima I, Yasutomo K, A mutation in the immunoproteasome subunit PSMB8 causes autoinflammation and lipodystrophy in humans. J. Clin. Invest 121, 4150–4160 (2011). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

