A computationally tractable birth-death model that combines phylogenetic and epidemiological data
- PMID: 35148311
- PMCID: PMC8903285
- DOI: 10.1371/journal.pcbi.1009805
A computationally tractable birth-death model that combines phylogenetic and epidemiological data
Abstract
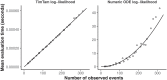
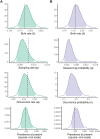
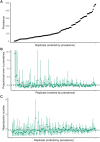
Inferring the dynamics of pathogen transmission during an outbreak is an important problem in infectious disease epidemiology. In mathematical epidemiology, estimates are often informed by time series of confirmed cases, while in phylodynamics genetic sequences of the pathogen, sampled through time, are the primary data source. Each type of data provides different, and potentially complementary, insight. Recent studies have recognised that combining data sources can improve estimates of the transmission rate and the number of infected individuals. However, inference methods are typically highly specialised and field-specific and are either computationally prohibitive or require intensive simulation, limiting their real-time utility. We present a novel birth-death phylogenetic model and derive a tractable analytic approximation of its likelihood, the computational complexity of which is linear in the size of the dataset. This approach combines epidemiological and phylodynamic data to produce estimates of key parameters of transmission dynamics and the unobserved prevalence. Using simulated data, we show (a) that the approximation agrees well with existing methods, (b) validate the claim of linear complexity and (c) explore robustness to model misspecification. This approximation facilitates inference on large datasets, which is increasingly important as large genomic sequence datasets become commonplace.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Brauer F, van den Driessche P, Wu J. Mathematical Epidemiology. Springer; 2008.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

