Rhizosphere bacteriome structure and functions
- PMID: 35149704
- PMCID: PMC8837802
- DOI: 10.1038/s41467-022-28448-9
Rhizosphere bacteriome structure and functions
Abstract
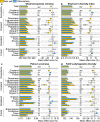
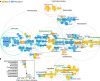
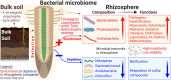
Microbial composition and functions in the rhizosphere-an important microbial hotspot-are among the most fascinating yet elusive topics in microbial ecology. We used 557 pairs of published 16S rDNA amplicon sequences from the bulk soils and rhizosphere in different ecosystems around the world to generalize bacterial characteristics with respect to community diversity, composition, and functions. The rhizosphere selects microorganisms from bulk soil to function as a seed bank, reducing microbial diversity. The rhizosphere is enriched in Bacteroidetes, Proteobacteria, and other copiotrophs. Highly modular but unstable bacterial networks in the rhizosphere (common for r-strategists) reflect the interactions and adaptations of microorganisms to dynamic conditions. Dormancy strategies in the rhizosphere are dominated by toxin-antitoxin systems, while sporulation is common in bulk soils. Functional predictions showed that genes involved in organic compound conversion, nitrogen fixation, and denitrification were strongly enriched in the rhizosphere (11-182%), while genes involved in nitrification were strongly depleted.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013;64:807–838. - PubMed
-
- Leach JE, Triplett LR, Argueso CT, Trivedi P. Communication in the phytobiome. Cell. 2017;169:587–596. - PubMed
-
- Vorholt JA, Vogel C, Carlstrom CI, Muller DB. Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe. 2017;22:142–155. - PubMed
-
- Jiang Y, et al. Plant cultivars imprint the rhizosphere bacterial community composition and association networks. Soil Biol. Biochem. 2017;109:145–155.
-
- Garbeva P, van Elsas JD, van Veen JA. Rhizosphere microbial community and its response to plant species and soil history. Plant Soil. 2008;302:19–32.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

