The spatial transcriptomic landscape of the healing mouse intestine following damage
- PMID: 35149721
- PMCID: PMC8837647
- DOI: 10.1038/s41467-022-28497-0
The spatial transcriptomic landscape of the healing mouse intestine following damage
Abstract
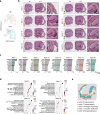
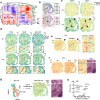
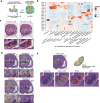
The intestinal barrier is composed of a complex cell network defining highly compartmentalized and specialized structures. Here, we use spatial transcriptomics to define how the transcriptomic landscape is spatially organized in the steady state and healing murine colon. At steady state conditions, we demonstrate a previously unappreciated molecular regionalization of the colon, which dramatically changes during mucosal healing. Here, we identified spatially-organized transcriptional programs defining compartmentalized mucosal healing, and regions with dominant wired pathways. Furthermore, we showed that decreased p53 activation defined areas with increased presence of proliferating epithelial stem cells. Finally, we mapped transcriptomics modules associated with human diseases demonstrating the translational potential of our dataset. Overall, we provide a publicly available resource defining principles of transcriptomic regionalization of the colon during mucosal healing and a framework to develop and progress further hypotheses.
© 2022. The Author(s).
Conflict of interest statement
E.J.V. and N.G. have received research grants from F. Hoffmann-La Roche. C.E., L.L. and J.L. are scientific consultants for 10X Genomics Inc. The remaining authors declare no competing interests.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

