Relationship between oxidative stress and lifespan in Daphnia pulex
- PMID: 35149730
- PMCID: PMC8837783
- DOI: 10.1038/s41598-022-06279-4
Relationship between oxidative stress and lifespan in Daphnia pulex
Abstract
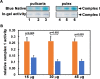
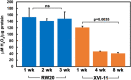
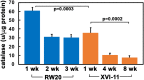
Macromolecular damage leading to cell, tissue and ultimately organ dysfunction is a major contributor to aging. Intracellular reactive oxygen species (ROS) resulting from normal metabolism cause most damage to macromolecules and the mitochondria play a central role in this process as they are the principle source of ROS. The relationship between naturally occurring variations in the mitochondrial (MT) genomes leading to correspondingly less or more ROS and macromolecular damage that changes the rate of aging associated organismal decline remains relatively unexplored. MT complex I, a component of the electron transport chain (ETC), is a key source of ROS and the NADH dehydrogenase subunit 5 (ND5) is a highly conserved core protein of the subunits that constitute the backbone of complex I. Using Daphnia as a model organism, we explored if the naturally occurring sequence variations in ND5 correlate with a short or long lifespan. Our results indicate that the short-lived clones have ND5 variants that correlate with reduced complex I activity, increased oxidative damage, and heightened expression of ROS scavenger enzymes. Daphnia offers a unique opportunity to investigate the association between inherited variations in components of complex I and ROS generation which affects the rate of aging and lifespan.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

