Formation and function of OmpG or OmpA-incorporated liposomes using an in vitro translation system
- PMID: 35149747
- PMCID: PMC8837779
- DOI: 10.1038/s41598-022-06314-4
Formation and function of OmpG or OmpA-incorporated liposomes using an in vitro translation system
Abstract
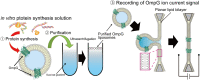
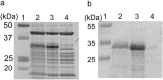
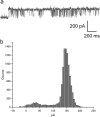
Outer membrane proteins (OMPs), located on the outer membrane of gram-negative bacteria, have a β-strand structure and form nanopores, which allow passage of ions, sugars, and small molecules. Recently, OMPs have been used as sensing elements to detect biological molecules. OMPs are normally expressed and purified from Escherichia coli (E. coli). Although the cell-free synthesis of OMPs, such as OmpA and OmpG, is achieved in the presence of liposomes and periplasmic chaperones, the amount of OmpA and OmpG incorporated into the nano-sized liposomes is not clear. In this study, after in vitro translation, the incorporation of OmpG into purified nano-sized liposomes with various lipid compositions was investigated. Liposomes containing the synthesized OmpG were purified using a stepwise sucrose density gradient. We report that liposomes prepared with the E. coli lipid extract (PE/PG) had the highest amount of OmpG incorporated compared to liposomes with other lipid compositions, as detected by recording the current across the OmpG containing liposomes using the patch clamp technique. This study reveals some of the requirements for the insertion and refolding of OMPs synthesized by the in vitro translation system into lipid membranes, which plays a role in the biological sensing of various molecules.
© 2022. The Author(s).
Conflict of interest statement
The author declares no competing interests.
Figures

Similar articles
-
Incorporation of outer membrane protein OmpG in lipid membranes: protein-lipid interactions and beta-barrel orientation.Biochemistry. 2008 Jun 10;47(23):6189-98. doi: 10.1021/bi800203g. Epub 2008 May 13. Biochemistry. 2008. PMID: 18473482
-
The Bam complex catalyzes efficient insertion of bacterial outer membrane proteins into membrane vesicles of variable lipid composition.J Biol Chem. 2018 Feb 23;293(8):2959-2973. doi: 10.1074/jbc.RA117.000349. Epub 2018 Jan 8. J Biol Chem. 2018. PMID: 29311257 Free PMC article.
-
Purification, Refolding, and Crystallization of the Outer Membrane Protein OmpG from Escherichia coli.Methods Enzymol. 2015;557:149-66. doi: 10.1016/bs.mie.2015.01.018. Epub 2015 Mar 24. Methods Enzymol. 2015. PMID: 25950964
-
Folding kinetics of the outer membrane proteins OmpA and FomA into phospholipid bilayers.Chem Phys Lipids. 2006 Jun;141(1-2):30-47. doi: 10.1016/j.chemphyslip.2006.02.004. Epub 2006 Mar 20. Chem Phys Lipids. 2006. PMID: 16581049 Review.
-
From Chaperones to the Membrane with a BAM!Trends Biochem Sci. 2016 Oct;41(10):872-882. doi: 10.1016/j.tibs.2016.06.005. Epub 2016 Jul 19. Trends Biochem Sci. 2016. PMID: 27450425 Free PMC article. Review.
Cited by
-
Cell-Penetrating Peptide-Mediated Biomolecule Transportation in Artificial Lipid Vesicles and Living Cells.Molecules. 2024 Jul 16;29(14):3339. doi: 10.3390/molecules29143339. Molecules. 2024. PMID: 39064917 Free PMC article. Review.
-
Bioengineered bacterial outer membrane vesicles encapsulated Polybia-mastoparan I fusion peptide as a promising nanoplatform for bladder cancer immune-modulatory chemotherapy.Front Immunol. 2023 Mar 14;14:1129771. doi: 10.3389/fimmu.2023.1129771. eCollection 2023. Front Immunol. 2023. PMID: 36999028 Free PMC article.
-
Efficiency of transcription and translation of cell-free protein synthesis systems in cell-sized lipid vesicles with changing lipid composition determined by fluorescence measurements.Sci Rep. 2024 Feb 3;14(1):2852. doi: 10.1038/s41598-024-53135-8. Sci Rep. 2024. PMID: 38310141 Free PMC article.
-
Cell-Free Expression of De Novo Designed Peptides That Form β-Barrel Nanopores.ACS Nano. 2023 Feb 28;17(4):3358-3367. doi: 10.1021/acsnano.2c07970. Epub 2023 Feb 2. ACS Nano. 2023. PMID: 36731872 Free PMC article.
-
Formation of Cell-Sized Liposomes Incorporating a β-Barrel-Structured Porin through Rehydration of a Phospholipid-Membrane Protein Dried Film.ACS Omega. 2024 Jan 26;9(5):5911-5918. doi: 10.1021/acsomega.3c09431. eCollection 2024 Feb 6. ACS Omega. 2024. PMID: 38343955 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

