Inducible directed evolution of complex phenotypes in bacteria
- PMID: 35150576
- PMCID: PMC9177967
- DOI: 10.1093/nar/gkac094
Inducible directed evolution of complex phenotypes in bacteria
Abstract
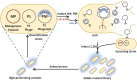
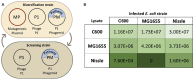
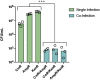
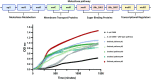
Directed evolution is a powerful method for engineering biology in the absence of detailed sequence-function relationships. To enable directed evolution of complex phenotypes encoded by multigene pathways, we require large library sizes for DNA sequences >5-10 kb in length, elimination of genomic hitchhiker mutations, and decoupling of diversification and screening steps. To meet these challenges, we developed Inducible Directed Evolution (IDE), which uses a temperate bacteriophage to package large plasmids and transfer them to naive cells after intracellular mutagenesis. To demonstrate IDE, we evolved a 5-gene pathway from Bacillus licheniformis that accelerates tagatose catabolism in Escherichia coli, resulting in clones with 65% shorter lag times during growth on tagatose after only two rounds of evolution. Next, we evolved a 15.4 kb, 10-gene pathway from Bifidobacterium breve UC2003 that aids E. coli's utilization of melezitose. After three rounds of IDE, we isolated evolved pathways that both reduced lag time by more than 2-fold and enabled 150% higher final optical density. Taken together, this work enhances the capacity and utility of a whole pathway directed evolution approach in E. coli.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
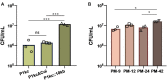


References
-
- Harvey A.L., Edrada-Ebel R., Quinn R.J.. The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 2015; 14:111–129. - PubMed
-
- Crook N., Alper H.S.. Patnaik R. Classical strain improvement. Engineering Complex Phenotypes in Industrial Strains. 2012;
-
- Lee S.Y., Kim H.U., Chae T.U., Cho J.S., Kim J.W., Shin J.H., Kim D.I., Ko Y.-S., Jang W.D., Jang Y.-S.. A comprehensive metabolic map for production of bio-based chemicals. Nat. Catal. 2019; 2:18–33.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

