RNA Binding Motif 5 Gene Deletion Modulates Cell Signaling in a Sex-Dependent Manner but Not Hippocampal Cell Death
- PMID: 35152732
- PMCID: PMC8978574
- DOI: 10.1089/neu.2021.0362
RNA Binding Motif 5 Gene Deletion Modulates Cell Signaling in a Sex-Dependent Manner but Not Hippocampal Cell Death
Abstract
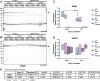
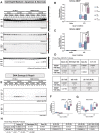
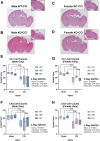
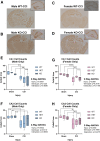
RNA-binding motif 5 (RBM5) is a pro-death tumor suppressor gene in cancer cells. It remains to be determined if it is neurotoxic in the brain or rather if it plays a fundamentally different role in the central nervous system (CNS). Brain-specific RBM5 knockout (KO) mice were given a controlled cortical impact (CCI) traumatic brain injury (TBI). Markers of acute cellular damage and repair were measured in hippocampal homogenates 48 h post-CCI. Hippocampal CA1/CA3 cell counts were assessed 7 days post-CCI to determine if early changes in injury markers were associated with histological outcome. No genotype-dependent differences were found in the levels of apoptotic markers (caspase 3, caspase 6, and caspase 9). However, KO females had a paradoxical increase in markers of pro-death calpain activation (145/150-spectrin and breakdown products [SBDP]) and in DNA repair/survival markers. (pH2A.x and pCREB). CCI-injured male KOs had a significant increase in phosphorylated calcium/calmodulin-dependent protein kinase II (pCaMKII). Despite sex/genotype-dependent differences in KOs in the levels of acute cell signaling targets involved in cell death pathways, 7 day hippocampal neuronal survival did not differ from that of wild types (WTs). Similarly, no differences in astrogliosis were observed. Finally, gene analysis revealed increased estrogen receptor α (ERα) levels in the KO hippocampus in females and may suggest a novel mechanism to explain sex-dimorphic effects on cell signaling. In summary, RBM5 inhibition did not affect hippocampal survival after a TBI in vivo but did modify targets involved in neural signal transduction/Ca2+ signaling pathways. Findings here support the view that RBM5 may serve a purpose in the CNS that is dissimilar from its traditional pro-death role in cancer.
Keywords: RBM5; RNA binding protein; gender; sexual dimorphism; trauma.
Conflict of interest statement
T.C.J. and P.M.K. are inventors of a USPTO patent (No. 9610266) titled: Small molecule inhibitors of RNA binding motif (RBM) proteins for the treatment of acute cellular injury. The other authors have nothing to disclose
Figures



References
-
- Oh, J.J., Razfar, A., Delgado, I., Reed, R.A., Malkina, A., Boctor, B., and Slamon, D.J. (2006). 3p21.3 tumor suppressor gene H37/Luca15/RBM5 inhibits growth of human lung cancer cells through cell cycle arrest and apoptosis. Cancer Res. 66, 3419–3427. - PubMed
-
- Kobayashi, T., Ishida, J., Musashi, M., Ota, S., Yoshida, T., Shimizu, Y., Chuma, M., Kawakami, H., Asaka, M., Tanaka, J., Imamura, M., Kobayashi, M., Itoh, H., Edamatsu, H., Sutherland, L.C., and Brachmann, R.K. (2011). p53 transactivation is involved in the antiproliferative activity of the putative tumor suppressor RBM5. Int. J. Cancer 128, 304–318. - PubMed
-
- Rintala-Maki, N.D., and Sutherland, L.C. (2004). LUCA-15/RBM5, a putative tumour suppressor, enhances multiple receptor-initiated death signals. Apoptosis 9, 475–484. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

