The curse of the uncultured fungus
- PMID: 35153529
- PMCID: PMC8828591
- DOI: 10.3897/mycokeys.86.76053
The curse of the uncultured fungus
Abstract
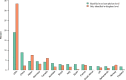
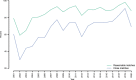
The international DNA sequence databases abound in fungal sequences not annotated beyond the kingdom level, typically bearing names such as "uncultured fungus". These sequences beget low-resolution mycological results and invite further deposition of similarly poorly annotated entries. What do these sequences represent? This study uses a 767,918-sequence corpus of public full-length fungal ITS sequences to estimate what proportion of the 95,055 "uncultured fungus" sequences that represent truly unidentifiable fungal taxa - and what proportion of them that would have been straightforward to annotate to some more meaningful taxonomic level at the time of sequence deposition. Our results suggest that more than 70% of these sequences would have been trivial to identify to at least the order/family level at the time of sequence deposition, hinting that factors other than poor availability of relevant reference sequences explain the low-resolution names. We speculate that researchers' perceived lack of time and lack of insight into the ramifications of this problem are the main explanations for the low-resolution names. We were surprised to find that more than a fifth of these sequences seem to have been deposited by mycologists rather than researchers unfamiliar with the consequences of poorly annotated fungal sequences in molecular repositories. The proportion of these needlessly poorly annotated sequences does not decline over time, suggesting that this problem must not be left unchecked.
Keywords: DNA barcoding; Data interoperability; data mining; scientific practice; species identification; taxonomic annotation.
Kessy Abarenkov, Erik Kristiansson, Martin Ryberg, Sandra Nogal-Prata, Daniela Gómez-Martínez, Katrin Stüer-Patowsky, Tobias Jansson, Sergei Põlme, Masoomeh Ghobad-Nejhad, Natàlia Corcoll, Ruud Scharn, Marisol Sánchez-García, Maryia Khomich, Christian Wurzbacher, R. Henrik Nilsson.
Figures


References
-
- Abarenkov K, Adams RI, Laszlo I, Agan A, Ambrosio E, Antonelli A, Bahram M, Bengtsson-Palme J, Bok G, Cangren P, Coimbra V, Coleine C, Gustafsson C, He J, Hofmann T, Kristiansson E, Larsson E, Larsson T, Liu Y, Martinsson S, Meyer W, Panova M, Pombubpa N, Ritter C, Ryberg M, Svantesson S, Scharn R, Svensson O, Töpel M, Unterseher M, Visagie C, Wurzbacher C, Taylor AFS, Kõljalg U, Schriml L, Nilsson RH. (2016) Annotating public fungal ITS sequences from the built environment according to the MIxS-Built Environment standard – a report from a May 23–24, 2016 workshop (Gothenburg, Sweden). MycoKeys 16: 1–15. 10.3897/mycokeys.16.10000 - DOI
-
- Baldrian P, Větrovský T, Lepinay C, Kohout P. (2021) High-throughput sequencing view on the magnitude of global fungal diversity. Fungal Diversity. 10.1007/s13225-021-00472-y - DOI
-
- Bengtsson‐Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A, DeWit P, Sanchez-Garcia M, Ebersberger I, de Sousa F, Amend AS, Jumpponen A, Unterseher M, Kristiansson E, Abarenkov K, Bertrand YJK, Sanli K, Eriksson KM, Vik U, Veldre V, Nilsson RH. (2013) Improved software detection and extraction of ITS1 and ITS 2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods in Ecology and Evolution 4(10): 914–919. 10.1111/2041-210X.12073 - DOI
LinkOut - more resources
Full Text Sources
