Comparative Analysis of the MADS-Box Genes Revealed Their Potential Functions for Flower and Fruit Development in Longan (Dimocarpus longan)
- PMID: 35154209
- PMCID: PMC8829350
- DOI: 10.3389/fpls.2021.813798
Comparative Analysis of the MADS-Box Genes Revealed Their Potential Functions for Flower and Fruit Development in Longan (Dimocarpus longan)
Abstract
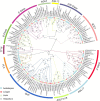
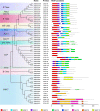
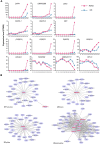
Longan (Dimocarpus longan Lour.) is an important economic crop widely planted in tropical and subtropical regions, and flower and fruit development play decisive effects on the longan yield and fruit quality formation. MCM1, AGAMOUS, DEFICIENS, Serum Response Factor (MADS)-box transcription factor family plays important roles for the flowering time, floral organ identity, and fruit development in plants. However, there is no systematic information of MADS-box family in longan. In this study, 114 MADS-box genes were identified from the longan genome, phylogenetic analysis divided them into type I (Mα, Mβ, Mγ) and type II (MIKC*, MIKC C ) groups, and MIKC C genes were further clustered into 12 subfamilies. Comparative genomic analysis of 12 representative plant species revealed the conservation of type II in Sapindaceae and analysis of cis-elements revealed that Dof transcription factors might directly regulate the MIKC C genes. An ABCDE model was proposed for longan based on the phylogenetic analysis and expression patterns of MADS-box genes. Transcriptome analysis revealed that MIKC C genes showed wide expression spectrums, particularly in reproductive organs. From 35 days after KClO3 treatment, 11 MIKC genes were up-regulated, suggesting a crucial role in off-season flower induction, while DlFLC, DlSOC1, DlSVP, and DlSVP-LIKE may act as the inhibitors. The gene expression patterns of longan fruit development indicated that DlSTK, DlSEP1/2, and DlMADS53 could be involved in fruit growth and ripening. This paper carried out the whole genome identification and analysis of the longan MADS-box family for the first time, which provides new insights for further understanding its function in flowers and fruit.
Keywords: ABCDE model; KClO3; MADS-box; flower; fruit development; longan.
Copyright © 2022 Wang, Hu, Fang, Feng, Fang, Zou, Zheng, Ming and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Characteristics of banana B genome MADS-box family demonstrate their roles in fruit development, ripening, and stress.Sci Rep. 2020 Nov 30;10(1):20840. doi: 10.1038/s41598-020-77870-w. Sci Rep. 2020. PMID: 33257717 Free PMC article.
-
Identification and Characterization of MIKCc-Type MADS-Box Genes in the Flower Organs of Adonis amurensis.Int J Mol Sci. 2021 Aug 28;22(17):9362. doi: 10.3390/ijms22179362. Int J Mol Sci. 2021. PMID: 34502271 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of the R2R3-MYB Transcription Factor Family Revealed Their Potential Roles in the Flowering Process in Longan (Dimocarpus longan).Front Plant Sci. 2022 Mar 25;13:820439. doi: 10.3389/fpls.2022.820439. eCollection 2022. Front Plant Sci. 2022. PMID: 35401601 Free PMC article.
-
The major clades of MADS-box genes and their role in the development and evolution of flowering plants.Mol Phylogenet Evol. 2003 Dec;29(3):464-89. doi: 10.1016/s1055-7903(03)00207-0. Mol Phylogenet Evol. 2003. PMID: 14615187 Review.
-
Genome-wide Analysis of the MADS-Box Gene Family in Watermelon.Comput Biol Chem. 2019 Jun;80:341-350. doi: 10.1016/j.compbiolchem.2019.04.013. Epub 2019 May 2. Comput Biol Chem. 2019. PMID: 31082717 Review.
Cited by
-
Genome-wide analysis of the MADS-box gene family of sea buckthorn (Hippophae rhamnoides ssp. sinensis) and their potential role in floral organ development.Front Plant Sci. 2024 Jun 13;15:1387613. doi: 10.3389/fpls.2024.1387613. eCollection 2024. Front Plant Sci. 2024. PMID: 38938643 Free PMC article.
-
Genome-wide identification and bioinformatics analysis of the WRKY transcription factors and screening of candidate genes for anthocyanin biosynthesis in azalea (Rhododendron simsii).Front Genet. 2023 May 10;14:1172321. doi: 10.3389/fgene.2023.1172321. eCollection 2023. Front Genet. 2023. PMID: 37234867 Free PMC article.
-
Unregulated GmAGL82 due to Phosphorus Deficiency Positively Regulates Root Nodule Growth in Soybean.Int J Mol Sci. 2024 Feb 1;25(3):1802. doi: 10.3390/ijms25031802. Int J Mol Sci. 2024. PMID: 38339080 Free PMC article.
-
Characterization of the MIKCC-type MADS-box gene family in blueberry and its possible mechanism for regulating flowering in response to the chilling requirement.Planta. 2024 Feb 29;259(4):77. doi: 10.1007/s00425-024-04349-7. Planta. 2024. PMID: 38421445
-
Genome-wide identification of the AcMADS-box family and functional validation of AcMADS32 involved in carotenoid biosynthesis in Actinidia.Front Plant Sci. 2023 Jun 19;14:1159942. doi: 10.3389/fpls.2023.1159942. eCollection 2023. Front Plant Sci. 2023. PMID: 37404538 Free PMC article.
References
-
- Aono N., Hasebe M. (2006). Functional analysis of MADS-box genes in Physcomitrella patens. Plant Cell Physiol. Suppl. 2006 498–498. 10.14841/jspp.2006.0.498.0 - DOI
-
- Bangerth K. F. (2009). Floral induction in mature, perennial angiosperm fruit trees: similarities and discrepancies with annual/biennial plants and the involvement of plant hormones. Sci. Hortic. 122 153–163. 10.1016/j.scienta.2009.06.014 - DOI
LinkOut - more resources
Full Text Sources

