Overcoming resistance to EGFR monotherapy in HNSCC by identification and inhibition of individualized cancer processes
- PMID: 35154483
- PMCID: PMC8771558
- DOI: 10.7150/thno.64347
Overcoming resistance to EGFR monotherapy in HNSCC by identification and inhibition of individualized cancer processes
Abstract
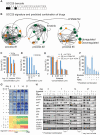
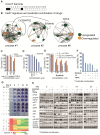
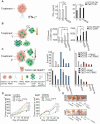
Therapeutic strategies for advanced head and neck squamous carcinoma (HNSCC) consist of multimodal treatment, including Epidermal Growth Factor Receptor (EGFR) inhibition, immune-checkpoint inhibition, and radio (chemo) therapy. Although over 90% of HNSCC tumors overexpress EGFR, attempts to replace cytotoxic treatments with anti-EGFR agents have failed due to alternative signaling pathways and inter-tumor heterogeneity. Methods: Using protein expression data obtained from hundreds of HNSCC tissues and cell lines we compute individualized signaling signatures using an information-theoretic approach. The approach maps each HNSCC malignancy according to the protein-protein network reorganization in every tumor. We show that each patient-specific signaling signature (PaSSS) includes several distinct altered signaling subnetworks. Based on the resolved PaSSSs we design personalized drug combinations. Results: We show that simultaneous targeting of central hub proteins from each altered subnetwork is essential to selectively enhance the response of HNSCC tumors to anti-EGFR therapy and inhibit tumor growth. Furthermore, we demonstrate that the PaSSS-based drug combinations lead to induced expression of T cell markers and IFN-γ secretion, pointing to higher efficiency of the immune response. Conclusion: The PaSSS-based approach advances our understanding of how individualized therapies should be tailored to HNSCC tumors.
Keywords: head and neck squamous cell carcinoma; information-theoretic approach; patient-specific signaling signatures; precision medicine; targeted therapy.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
-
- Kareemaghay S, Tavassoli M. Clinical immunotherapeutic approaches for the treatment of head and neck cancer. Int J Oral Maxillofac Surg. 2019;48:419–36. - PubMed
-
- Ribeiro FAP, Noguti J, Oshima CTF, Ribeiro DA. Effective targeting of the epidermal growth factor receptor (EGFR) for treating oral cancer: a promising approach. Anticancer Res. 2014;34:1547–52. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

