Engineering Crystal Packing in RNA-Protein Complexes II: A Historical Perspective from the Structural Studies of the Spliceosome
- PMID: 35154816
- PMCID: PMC7612351
- DOI: 10.3390/cryst11080948
Engineering Crystal Packing in RNA-Protein Complexes II: A Historical Perspective from the Structural Studies of the Spliceosome
Abstract
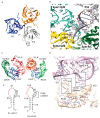
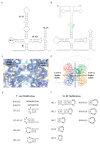
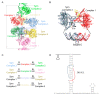
Cryo-electron microscopy has greatly advanced our understanding of how the spliceosome cycles through different conformational states to conduct the chemical reactions that remove introns from pre-mRNA transcripts. The Cryo-EM structures were built upon decades of crystallographic studies of various spliceosomal RNA-protein complexes. In this review we give an overview of the crystal structures solved in the Nagai group, utilizing many of the strategies to design crystal packing as described in the accompanying paper.
Keywords: RNA-protein complexes; crystallization; spliceosome.
Conflict of interest statement
Conflicts of Interest: The authors declare no conflict of interest.
Figures



References
-
- Crowther RA. In: Methods in Enzymology. Crowther RA, editor. Vol. 579. Academic Press; Cambridge, MA, USA: 2016. Preface; pp. xiii–xx. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
