Analysis of miRNAs Involved in Mouse Heart Injury Upon Coxsackievirus A2 Infection
- PMID: 35155276
- PMCID: PMC8831793
- DOI: 10.3389/fcimb.2022.765445
Analysis of miRNAs Involved in Mouse Heart Injury Upon Coxsackievirus A2 Infection
Abstract
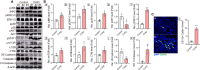
Coxsackievirus A2 (CVA2) has recently been constantly detected, and is associated with viral myocarditis in children. Our previous study demonstrated that CVA2 led to heart damage in a neonatal murine model. However, the molecular mechanism of heart injury caused by CVA2 remains largely unknown. Emerging evidence suggests the significant functions of miRNAs in Coxsackievirus infection. To investigate potential miRNAs involved in heart injury caused by CVA2, our study, for the first time, conducted a RNA-seq in vivo employing infected mice hearts. In total, 87, 101 and 76 differentially expressed miRNAs were identified at 3 days post infection (dpi), 7 dpi and 7 dpi vs 3 dpi. Importantly, above 3 comparison strategies shared 34 differentially expressed miRNAs. These results were confirmed by quantitative PCR (qPCR). Next, we did GO, KEGG, and miRNA-mRNA integrated analysis of differential miRNAs. The dual-luciferase reporter assay confirmed the miRNA-mRNA pairs. To further confirm the above enriched pathways and processes, we did Western blotting and immunofluorescence staining. Our results suggest that inflammatory responses, T cell activation, apoptosis, autophagy, antiviral immunity, NK cell infiltration, and the disruption of tight junctions are involved in the pathogenesis of heart injury caused by CVA2. The dysregulated miRNAs and pathways recognized in the current study can improve the understanding of the intricate interactions between CVA2 and the heart injury, opening a novel avenue for the future study of CVA2 pathogenesis.
Keywords: Coxsackievirus A2; RNA-seq; and mouth disease; foot; hand; miRNAs; molecular mechanism.
Copyright © 2022 Wu, Zhu, Qian, Hu, Ji, Li, Zhu, Liang and Jin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Coxsackievirus A2 Leads to Heart Injury in a Neonatal Mouse Model.Viruses. 2021 Aug 11;13(8):1588. doi: 10.3390/v13081588. Viruses. 2021. PMID: 34452454 Free PMC article.
-
Analysis of miRNAs involved in mouse brain injury upon Coxsackievirus A6 infection.Front Cell Infect Microbiol. 2024 Aug 22;14:1405689. doi: 10.3389/fcimb.2024.1405689. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39239635 Free PMC article.
-
The CXCL10/CXCR3 Axis Promotes Disease Pathogenesis in Mice upon CVA2 Infection.Microbiol Spectr. 2022 Jun 29;10(3):e0230721. doi: 10.1128/spectrum.02307-21. Epub 2022 May 23. Microbiol Spectr. 2022. PMID: 35604176 Free PMC article.
-
The Disruption of the Endothelial Barrier Contributes to Acute Lung Injury Induced by Coxsackievirus A2 Infection in Mice.Int J Mol Sci. 2021 Sep 13;22(18):9895. doi: 10.3390/ijms22189895. Int J Mol Sci. 2021. PMID: 34576058 Free PMC article.
-
RNA-Seq Revealed a Circular RNA-microRNA-mRNA Regulatory Network in Hantaan Virus Infection.Front Cell Infect Microbiol. 2020 Mar 13;10:97. doi: 10.3389/fcimb.2020.00097. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32232013 Free PMC article. Review.
Cited by
-
The key mechanisms of multi-system responses triggered by central nervous system damage in hand, foot, and mouth disease severity.Infect Med (Beijing). 2024 Jul 27;3(3):100124. doi: 10.1016/j.imj.2024.100124. eCollection 2024 Sep. Infect Med (Beijing). 2024. PMID: 39314804 Free PMC article. Review.
-
MicroRNAs in Myocarditis-Review of the Preclinical In Vivo Trials.Biomedicines. 2023 Oct 8;11(10):2723. doi: 10.3390/biomedicines11102723. Biomedicines. 2023. PMID: 37893097 Free PMC article. Review.
References
-
- Ai Y., Zhang W., Wu J., Zhang J., Shen M., Yao S., et al. . (2021). Molecular Epidemiology and Clinical Features of Enteroviruses-Associated Hand, Foot, and Mouth Disease and Herpangina Outbreak in Zunyi, China, 2019. Front. Med. (Lausanne) 8, 656699. doi: 10.3389/fmed.2021.656699 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

