Non-coding RNA and cholesteatoma
- PMID: 35155784
- PMCID: PMC8823169
- DOI: 10.1002/lio2.728
Non-coding RNA and cholesteatoma
Abstract
Objective: Cholesteatoma is a challenging chronic pathology of the middle ear for which pharmacologic therapies have not been developed yet. Cholesteatoma occurrence depends on the interplay between genetic and environmental factors while master regulators orchestrating disease progression are still unknown. Therefore, in this review, we will discuss the diagnostic and therapeutic potential of non-coding RNAs (ncRNA) as a new class of regulatory molecules.
Methods: We have comprehensively reviewed all articles investigating ncRNAs, specifically micro RNAs (miRNAs) and long ncRNAs (lncRNA/circRNA) in cholesteatoma tissue.
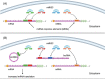
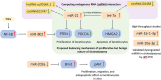
Results: Candidate miRNA approaches indicated that miR-21 and let-7a are the major miRNAs involved in cholesteatoma growth, migration, proliferation, bone destruction, and apoptosis. Regulatory potential for the same biological processes was also observed for miR-203a. The NF-kB/miR-802/PTEN regulatory network was in relation to observed miR-21 activity in cholesteatoma as well. High throughput approaches revealed additional ncRNAs implicated in cholesteatoma pathology. Competitive endogenous RNA (ceRNA) analysis highlighted lncRNA/circRNA that could be "endogenous sponge" for miR-21 and let-7a based on the hypothesis that RNA transcripts can communicate with and regulate each other by using shared miRNA response elements.
Conclusion: In this review, we summarize the discoveries and role of ncRNA in major pathways in cholesteatoma and highlight the potential of miRNA-based therapeutics in the treatment of cholesteatoma.
Level of Evidence: NA.
Keywords: RNA interaction; cholesteatoma; long noncoding RNA; micro RNA; noncoding RNA.
© 2022 The Authors. Laryngoscope Investigative Otolaryngology published by Wiley Periodicals LLC on behalf of The Triological Society.
Conflict of interest statement
The authors declare no potential conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
