Gene age shapes the transcriptional landscape of sexual morphogenesis in mushroom-forming fungi (Agaricomycetes)
- PMID: 35156613
- PMCID: PMC8893723
- DOI: 10.7554/eLife.71348
Gene age shapes the transcriptional landscape of sexual morphogenesis in mushroom-forming fungi (Agaricomycetes)
Abstract
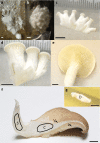
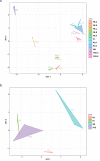
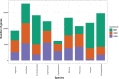
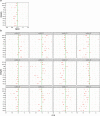
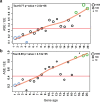
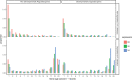
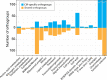
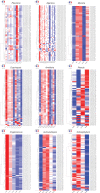
Multicellularity has been one of the most important innovations in the history of life. The role of gene regulatory changes in driving transitions to multicellularity is being increasingly recognized; however, factors influencing gene expression patterns are poorly known in many clades. Here, we compared the developmental transcriptomes of complex multicellular fruiting bodies of eight Agaricomycetes and Cryptococcus neoformans, a closely related human pathogen with a simple morphology. In-depth analysis in Pleurotus ostreatus revealed that allele-specific expression, natural antisense transcripts, and developmental gene expression, but not RNA editing or a 'developmental hourglass,' act in concert to shape its transcriptome during fruiting body development. We found that transcriptional patterns of genes strongly depend on their evolutionary ages. Young genes showed more developmental and allele-specific expression variation, possibly because of weaker evolutionary constraint, suggestive of nonadaptive expression variance in fruiting bodies. These results prompted us to define a set of conserved genes specifically regulated only during complex morphogenesis by excluding young genes and accounting for deeply conserved ones shared with species showing simple sexual development. Analysis of the resulting gene set revealed evolutionary and functional associations with complex multicellularity, which allowed us to speculate they are involved in complex multicellular morphogenesis of mushroom fruiting bodies.
Keywords: Coprinopsis cinerea; Cryptococcus neoformans; Pleurotus ostreatus; allelic imbalance; developmental biology; developmental hourglass; evolutionary biology; mushroom-forming fungi.
© 2022, Merényi et al.
Conflict of interest statement
ZM, MV, EG, JS, BK, TV, AG, BH, BB, LN No competing interests declared
Figures


















References
-
- Alexa A, Rahnenfuhrer J. Gene set enrichment analysis with topGO. Bioconductor. 2016. [February 4, 2019]. https://bioconductor.org/packages/release/bioc/vignettes/topGO/inst/doc/...
-
- Alfaro M, Castanera R, Lavín JL, Grigoriev IV, Oguiza JA, Ramírez L, Pisabarro AG. Comparative and transcriptional analysis of the predicted secretome in the lignocellulose-degrading basidiomycete fungus Pleurotus ostreatus. Environmental Microbiology. 2016;18:4710–4726. doi: 10.1111/1462-2920.13360. - DOI - PubMed
-
- Almási É, Sahu N, Krizsán K, Bálint B, Kovács GM, Kiss B, Cseklye J, Drula E, Henrissat B, Nagy I, Chovatia M, Adam C, LaButti K, Lipzen A, Riley R, Grigoriev IV, Nagy LG. Comparative genomics reveals unique wood-decay strategies and fruiting body development in the Schizophyllaceae. The New Phytologist. 2019;224:902–915. doi: 10.1111/nph.16032. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Dryad/10.5061/dryad.5qfttdz5m
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

