Distinct metabolic hallmarks of WHO classified adult glioma subtypes
- PMID: 35157758
- PMCID: PMC9435506
- DOI: 10.1093/neuonc/noac042
Distinct metabolic hallmarks of WHO classified adult glioma subtypes
Abstract
Background: Gliomas are complex tumors with several genetic aberrations and diverse metabolic programs contributing to their aggressive phenotypes and poor prognoses. This study defines key metabolic features that can be used to differentiate between glioma subtypes, with potential for improved diagnostics and subtype targeted therapy.
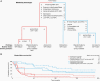
Methods: Cross-platform global metabolomic profiling coupled with clinical, genetic, and pathological analysis of glioma tissue from 224 tumors-oligodendroglioma (n = 31), astrocytoma (n = 31) and glioblastoma (n = 162)-were performed. Identified metabolic phenotypes were evaluated in accordance with the WHO classification, IDH-mutation, 1p/19q-codeletion, WHO-grading 2-4, and MGMT promoter methylation.
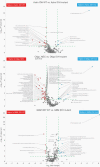
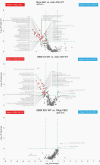
Results: Distinct metabolic phenotypes separate all six analyzed glioma subtypes. IDH-mutated subtypes, expressing 2-hydroxyglutaric acid, were clearly distinguished from IDH-wildtype subtypes. Considerable metabolic heterogeneity outside of the mutated IDH pathway were also evident, with key metabolites being high expression of glycerophosphates, inositols, monosaccharides, and sugar alcohols and low levels of sphingosine and lysoglycerophospholipids in IDH-mutants. Among the IDH-mutated subtypes, we observed high levels of amino acids, especially glycine and 2-aminoadipic acid, in grade 4 glioma, and N-acetyl aspartic acid in low-grade astrocytoma and oligodendroglioma. Both IDH-wildtype and mutated oligodendroglioma and glioblastoma were characterized by high levels of acylcarnitines, likely driven by rapid cell growth and hypoxic features. We found elevated levels of 5-HIAA in gliosarcoma and a subtype of oligodendroglioma not yet defined as a specific entity, indicating a previously not described role for the serotonin pathway linked to glioma with bimorphic tissue.
Conclusion: Key metabolic differences exist across adult glioma subtypes.
Keywords: WHO classification; astrocytoma; glioblastoma; metabolic reprogramming; oligodendroglioma.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Society for Neuro-Oncology.
Figures



Comment in
-
What can metabolites tell us about gliomas?Neuro Oncol. 2022 Sep 1;24(9):1469-1470. doi: 10.1093/neuonc/noac128. Neuro Oncol. 2022. PMID: 35554565 Free PMC article. No abstract available.
References
-
- Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

